Understanding how genetic information is processed within cells is crucial. One of the key steps in this process involves the maturation of pre-mRNAs (precursor messenger RNAs) in eukaryotic cells, which includes splicing and polyadenylation. This maturation is finely tuned by various RNA-binding proteins (RBPs). With over 1,500 RBPs present in human cells, scientists have a massive task in understanding how these proteins function and regulate RNA processing, especially within the complex environments of tissues and in the context of diseases.
The Role of RNA-Binding Proteins (RBPs)
RBPs are essential players in the processing of pre-mRNA, which is a preliminary form of mRNA that needs to be spliced and have a poly-A tail added before it can be translated into a protein. Splicing involves cutting out non-coding regions (introns) and joining coding regions (exons) together. Polyadenylation adds a tail of adenine nucleotides to the end of the mRNA, which protects it from degradation and aids in its export from the nucleus and translation into a protein.
Each RBP has specific binding motifs—sequences of RNA that they recognize and bind to. However, identifying these motifs and understanding the specific roles of RBPs has been challenging due to the complexity and variability of tissues and diseases.
Introducing MAPP: A New Tool for RNA Research
To address this challenge, scientists at the Swiss Institute of Bioinformatics have developed a new tool called MAPP (Motif Activity on Pre-mRNA Processing). MAPP is designed to systematically and automatically detect sequence motif-guided pre-mRNA processing regulation from RNA sequencing (RNA-Seq) data. This innovative tool allows researchers to explore how RBPs regulate RNA processing by analyzing the sequences they bind to and their effects on pre-mRNA.
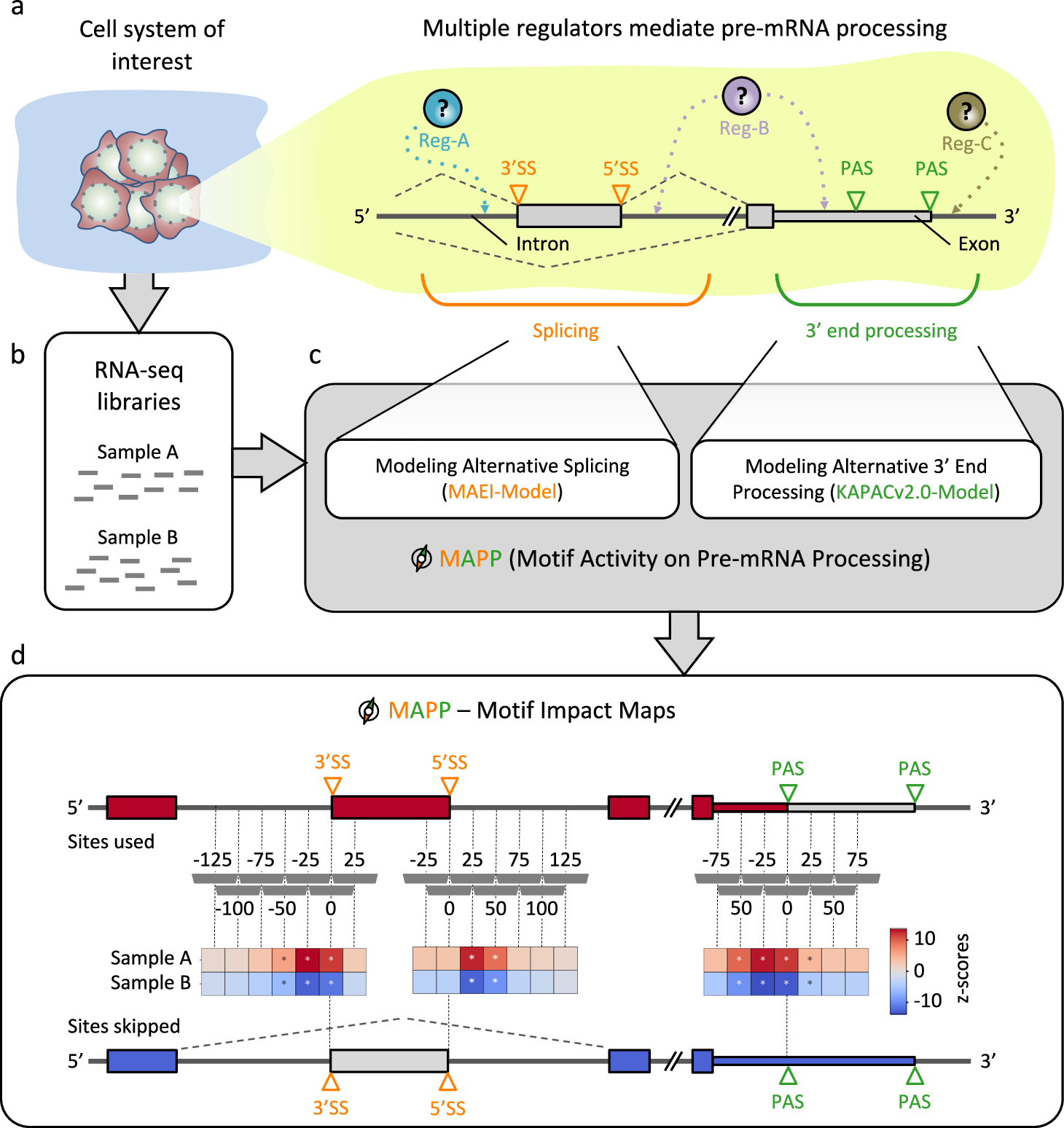
Inferring maps of RBP impact on splicing and 3’ end processing with MAPP
a Sketch illustrating how regulators (Reg) bind pre-mRNAs to influence the usage of splice sites (SS) and/or poly(A) sites (PAS). b RNA sequencing (RNA-seq) libraries are available or can be created for most cellular systems of interest. c MAPP analyzes the splicing and 3′ end processing patterns apparent in the RNA-Seq data with the MAEI (Motif Activity on Exon Inclusion) and KAPACv2.0 (K-mer Activity on PolyAdenylation site Choice version 2.0) models, respectively. d MAPP infers regulatory motifs for RBPs and reports detailed maps of their position-dependent impact on cassette exon inclusion and poly(A) site usage, respectively, by applying the models to genomic windows located at specific distances relative to the RNA processing sites (dashed gray vertical bars).
How MAPP Works and Its Discoveries
By applying MAPP to experiments where specific RBPs are knocked down (i.e., their activity is reduced), the researchers have discovered that many RBPs regulate both splicing and polyadenylation through similar sequence motifs. MAPP not only identifies these motifs but also reveals how the position of these motifs affects the role of RBPs in pre-mRNA processing.
Interestingly, the analysis showed that RBPs which influence both splicing and the 3′ end processing (polyadenylation) tend to have a consistent effect—either repressing or activating both processes. This consistency suggests that there might be an underlying mechanism that governs the dual roles of these RBPs.
MAPP’s Impact on Understanding Diseases
One of the most exciting applications of MAPP is in studying disease conditions. When applied to brain tissue samples from both healthy individuals and those with malignant brain tumors (glioblastomas), MAPP unveiled that RBPs like PTBP1 and RBFOX drive the oncogenic splicing program in glioblastomas. This means that these proteins contribute to the abnormal splicing patterns seen in cancer cells, which can promote tumor growth and survival.
The Future of RNA Research with MAPP
MAPP represents a significant advancement in the field of RNA biology. By providing a tool to systematically analyze the roles of RBPs in pre-mRNA processing, MAPP paves the way for a deeper understanding of how RNA processing is regulated in different conditions, both normal and pathological. This knowledge could eventually lead to new therapeutic strategies for diseases where RNA processing goes awry, such as cancer.
In summary, MAPP is an innovative tool that helps scientists decipher the complex roles of RNA-binding proteins in pre-mRNA maturation. By unveiling the sequence motifs and the regulatory mechanisms of RBPs, MAPP opens new avenues for understanding and potentially treating diseases linked to RNA processing.
Availability – The MAPP code is available on GitHub (https://github.com/gruber-sciencelab/MAPP)
Bak M, van Nimwegen E, Kouzel IU, Gur T, Schmidt R, Zavolan M, Gruber AJ. (2024) MAPP unravels frequent co-regulation of splicing and polyadenylation by RNA-binding proteins and their dysregulation in cancer. Nat Commun 15(1):4110. [article]