RNA has long been regarded as the messenger that carries genetic instructions from DNA to protein synthesis. However, recent advancements have uncovered a hidden layer of complexity within RNA known as the epitranscriptome. This complex landscape of RNA modifications, including methylation of adenine (m6A) and cytosine (m5C), plays a crucial role in regulating gene expression and cellular function.
Despite the growing interest in epitranscriptomics, researchers have faced a significant challenge: identifying multiple modifications within individual RNA molecules. Traditional methods have struggled to decipher this complex code, limiting our understanding of RNA’s diverse functions. However, researchers at the EMBL Australia Partner Laboratory Network have developed a groundbreaking new tool called CHEUI (CH3 Estimation Using Ionic current) that promises to overcome this obstacle and unlock the secrets of the epitranscriptome.
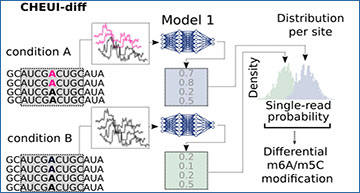
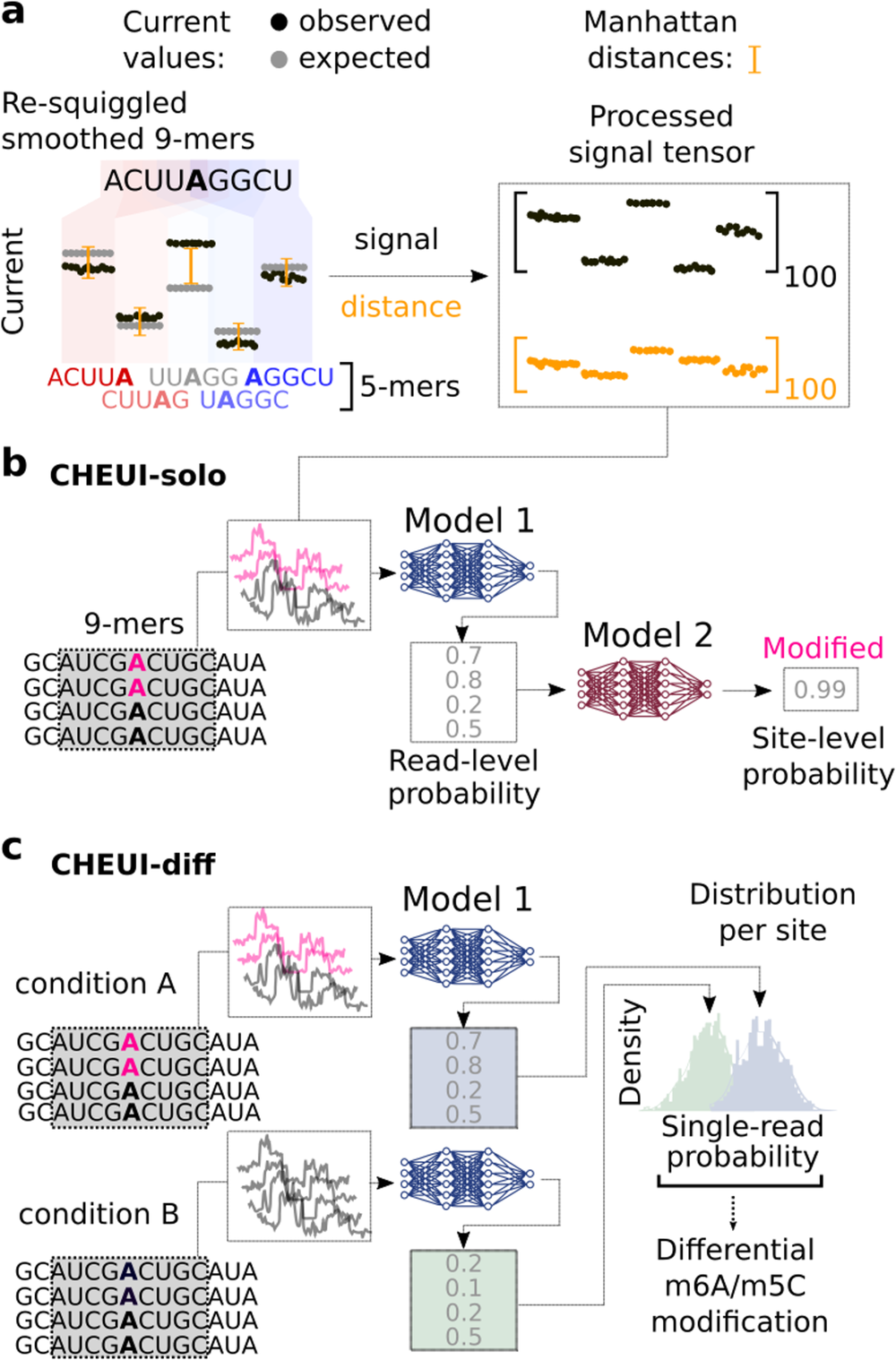
CHEUI architecture, modules, and signal processing approach
a CHEUI first processes signals for each 9 mer consisting of five consecutive overlapping 5 mers. The signals for each 5 mer are converted into 20 median values, yielding a vector of length 100. A vector of length 100 is obtained for the expected (unmodified) signals for the same five 5 mers and a vector of distances between the expected and observed signal values is calculated. These signal and distance vectors are used as inputs for Model 1. b CHEUI-solo Model 1 takes the signal and distance vectors corresponding to individual read signals associated with a 9 mer centered at every A (indicated as modified in pink, or unmodified in black) or C, and predicts the probability for each read of being modified A (m6A model) or modified C (m5C model). Model 2 uses the distribution of Model 1 probabilities for all the read signals at each reference transcript site and predicts the probability of the site being methylated and its stoichiometry, which estimated as the proportion of modified reads from Model 1 at that site. c CHEUI-diff uses the individual read probabilities from Model 1 in any two conditions to test for differential m6A or m5C at reference transcript sites using a two-tailed Mann–Whitney U-test.
CHEUI represents a significant leap forward in the field of epitranscriptomics by enabling the simultaneous detection of m6A and m5C modifications within individual RNA molecules. This innovative approach leverages nanopore direct RNA sequencing technology to analyze RNA at unprecedented resolution, providing insights into the stoichiometry and differential methylation between different conditions.
Key Features of CHEUI:
- Predicting Multiple Modifications: CHEUI utilizes advanced computational algorithms to predict the presence of both m6A and m5C modifications within the same RNA molecule. By analyzing nanopore sequencing signals, CHEUI achieves high accuracy in identifying these modifications, shedding light on the intricate landscape of the epitranscriptome.
- High Accuracy and Precision: In tests using synthetic RNA standards and real-world cell line data, CHEUI demonstrated remarkable accuracy and precision in predicting RNA modifications. Its ability to achieve high single-molecule, transcript-site, and stoichiometry accuracies sets a new standard in epitranscriptomic analysis.
- Revealing Co-Occurrence of Modifications: One of the most exciting findings enabled by CHEUI is the discovery of co-occurring m6A and m5C modifications within individual mRNA molecules. This revelation challenges previous assumptions about RNA modification patterns and opens up new avenues for exploring the functional significance of these modifications in cellular processes.
By enabling the simultaneous detection of multiple RNA modifications within individual molecules, CHEUI provides researchers with unprecedented insights into RNA’s hidden code. As scientists continue to unravel the mysteries of the epitranscriptome, tools like CHEUI will play a vital role in unlocking the secrets of gene regulation, cellular function, and disease mechanisms.
Availability – CHEUI is freely available from https://github.com/comprna/CHEUI under an Academic Public License.
Acera Mateos P, J Sethi A, Ravindran A, Srivastava A, Woodward K, Mahmud S, Kanchi M, Guarnacci M, Xu J, W S Yuen Z, Zhou Y, Sneddon A, Hamilton W, Gao J, M Starrs L, Hayashi R, Wickramasinghe V, Zarnack K, Preiss T, Burgio G, Dehorter N, E Shirokikh N, Eyras E. (2024) Prediction of m6A and m5C at single-molecule resolution reveals a transcriptome-wide co-occurrence of RNA modifications. Nat Commun 15(1):3899. [article]