Prioritization or ranking of different cell types in a single-cell RNA sequencing (scRNA-seq) framework can be performed in a variety of ways, some of these include: i) obtaining an indication of the proportion of cell types between the different conditions under study, ii) counting the number of differentially expressed genes (DEGs) between cell types and conditions in the experiment or, iii) prioritizing cell types based on prior knowledge about the conditions under study (i.e., a specific disease). These methods have drawbacks and limitations thus novel methods for improving cell ranking are required.
Researchers at the Cyprus Institute of Neurology & Genetics have developed a novel methodology that exploits prior knowledge in combination with expert-user information to accentuate cell types from a scRNA-seq analysis that yield the most biologically meaningful results with respect to a disease under study. This methodology allows for ranking and prioritization of cell types based on how well their expression profiles relate to the molecular mechanisms and drugs associated with a disease. Molecular mechanisms, as well as drugs, are incorporated as prior knowledge in a standardized, structured manner. Cell types are then ranked/prioritized based on how well results from data-driven analysis of scRNA-seq data match the predefined prior knowledge. In additional cell-cell communication perturbations between disease and control networks are used to further prioritize/rank cell types. This methodology has substantial advantages to more traditional cell ranking techniques and provides an informative complementary methodology that utilizes prior knowledge in a rapid and automated manner, that has previously not been attempted by other studies.
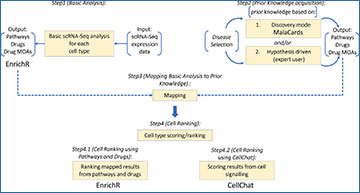
Flowchart of overall adopted methodology
Step 1 (Basic Analysis): Basic scRNA-seq analysis using SEURAT resulting in enriched pathways and repurposed drugs. Step 2 (Prior Knowledge Acquisition): Defining prior knowledge. Two options are available: i) proceed with all the terms obtained from a check list of predefined terms provided by querying MalaCards with a disease of interest ii) to perform a hypothesis-driven approach whereby the user can provide specific keywords/terms associated with the hypothesis under investigation to perform a de novo search across the supported databases. Step 3 (Mapping Basic Analysis to Prior Knowledge): Mapping step. Merges output from Steps 1 and 2 and assesses how well prior knowledge “maps” to the results obtained from scRNA-seq analysis. This is done firstly by mapping prior knowledge pathways against pathway enrichment results attained from analyzing the scRNA-seq data. Secondly, prior knowledge drug names and/or drug mode of actions (MOAs) are mapped against drug repurposing results from analyzing the scRNA-seq data using the CMAP database. Step 3 is performed for all cell types in the analysis. Step 4 (Cell Ranking): Scoring and ranking the cell types with respect to how well the data-driven output from pathway enrichment analysis and drug repurposing for individual cell types, “maps” to the predefined information provided by the expert user. Step 4 is further split into 2 steps (4.1 (Cell Ranking using Pathways and Drugs) and 4.2 (Cell Ranking using CellChat)): Step 4.1—Matching the position of the prior knowledge in the output (enriched pathways and repurposed drugs) of the scRNA-seq analysis and then taking the Euclidian distance between the matched positions. Step 4.2—Ranking of cells using cell-cell communication networks generated using CellChat. Performing a comparison of the number of interactions (edges) between cell types (nodes) in the two different networks (control vs. disease) and ranking the cells by log fold difference in interactions (LogFDI) taking in consideration both positive and negative fold changes. Finally, the union between results is obtained (denoted by U above) taking into consideration the top 3 ranked cell types from Steps 4.1 and 4.2.
Availability – The current methodology is also implemented as an R package entitled Single Cell Ranking Analysis Toolkit (scRANK) and is available for download and installation via GitHub (https://github.com/aoulas/scRANK).
ulas A, Savva K, Karathanasis N, Spyrou GM (2024) Ranking of cell clusters in a single-cell RNA-sequencing analysis framework using prior knowledge. PLoS Comput Biol 20(4): e1011550. [article]