Circular RNA (circRNA) has garnered considerable attention in recent years due to its regulatory roles in gene expression. While much research has focused on its interactions with microRNAs and proteins, little is known about its interactions with protein-coding mRNAs within intact cells. A groundbreaking study has now introduced a novel technology called Cross-Linking Poly(A) Pulldown RNase R Sequencing (CLiPPR-seq), shedding light on these elusive circRNA-mRNA hybrids.
Researchers at the DBT-Institute of Life Sciences employed AMT-mediated proximity ligation of RNA-RNA duplexes followed by circRNA enrichment and deep sequencing to develop the CLiPPR-seq method. This innovative approach identified hundreds of mRNA-interacting circRNAs across three different cell types: βTC6, C2C12, and HeLa cells. Additionally, CLiPP-seq without RNase R treatment was performed to assess mRNA expression in these cells, providing valuable insight into the overall gene expression landscape.
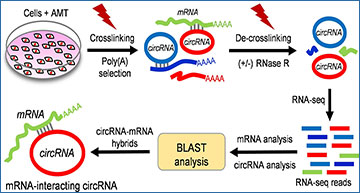
A step-by-step schematic representation of CLiPPR-Seq demonstrating the mapping of the circRNA-mRNA interactome by AMT crosslinking, poly(A) pulldown, RNase R treatment and sequencing to identify mRNA-interacting circRNAs.
Through BLAST analysis, circRNAs identified in CLiPPR-seq samples were cross-referenced with mRNAs from CLiPP-seq samples to identify potential complementary sequences for circRNA-mRNA interaction. Further confirmation of circRNA-mRNA interactions was obtained through pulldown assays of circRNAs and poly(A) RNAs, validating the direct interaction of circRNAs with target mRNAs.
One of the most intriguing findings of the study was the observation that silencing mRNA-interacting circRNAs led to altered expression of target mRNAs in βTC6 cells. This suggests a direct regulatory role of circRNAs in modulating gene expression, highlighting the functional significance of circRNA-mRNA interactions.
Overall, CLiPPR-seq represents a groundbreaking method for uncovering the complex network of circRNA-mRNA hybrids that may play critical roles in gene regulation. By illuminating these previously uncharacterized interactions, this technology opens new avenues for understanding the intricate mechanisms underlying gene expression regulation. Further research in this area holds the potential to unravel novel therapeutic targets and diagnostic biomarkers, ultimately advancing our knowledge of cellular biology and disease pathology.
Singh S, Shyamal S, Das A, Panda AC. (2024) Global identification of mRNA-interacting circular RNAs by CLiPPR-Seq. Nucleic Acids Res [Epub ahead of print]. [article]