The use of single cell sequencing technologies has exploded over recent years, and is now commonly used in many non-model species. Sequencing nuclei instead of whole cells has become increasingly popular, as it does not require the processing of samples immediately after collection. Researchers at The University of Edinburgh have developed a highly effective nucleus isolation protocol that outperforms previously available method in challenging samples in a non-model specie. This protocol can be successfully applied to extract nuclei from a variety of tissues and species.
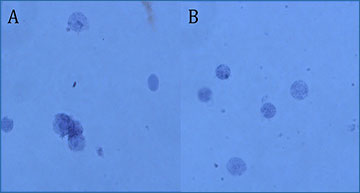
Three examples of nuclear dissociation trials in Atlantic salmon liver
at x40 magnification, stained with trypan blue
A. Nuclei did not spend sufficient time in the dissociation buffer resulting in incomplete dissociation. This may result in the obstruction of the microfluidic device and failure of the library. B. Nuclei perfectly dissociated showing minimal aggregation, ideal for library preparation. C. Nuclei have been in the dissociation buffer for too long, resulting in degradation of the nuclear membrane, which will result in suboptimal library quality.
Ruiz Daniels R, Taylor RS, Dobie R, Salisbury S, Furniss JJ, Clark E, et al. (2023) A versatile nuclei extraction protocol for single nucleus sequencing in non-model species–Optimization in various Atlantic salmon tissues. PLoS ONE 18(9): e0285020. [article]