Oxford Nanopore Technologies’ (ONT) sequencing platform offers an excellent opportunity to perform real-time analysis during sequencing. This feature allows for early insights into experimental data and accelerates a potential decision-making process for further analysis, which can be particularly relevant in the clinical context. Although some tools for the real-time analysis of DNA-sequencing data already exist, there is currently no application available for differential transcriptome data analysis designed for scientists or physicians with limited bioinformatics knowledge.
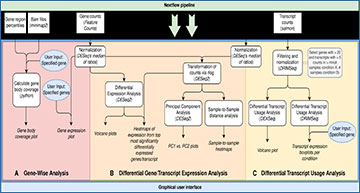
Researchers at the University Medical Center of the Johannes Gutenberg University have developed NanopoReaTA, a user-friendly real-time analysis toolbox for RNA sequencing data from ONT. Sequencing results from a running or finished experiment are processed through an R Shiny-based graphical user interface (GUI) with an integrated Nextflow pipeline for whole transcriptome or gene-specific analyses. NanopoReaTA provides visual snapshots of a sequencing run in progress, thus enabling interactive sequencing and rapid decision-making that could also be applied to clinical cases.
Availability – https://github.com/AnWiercze/NanopoReaTA and https://doi.org/10.5281/zenodo.8099825
Wierczeiko A, Pastore S, Mündnich S, Busch AM, Dietrich V, Helm M, Butto T, Gerber S. (2023) NanopoReaTA: a user-friendly tool for nanopore-seq real-time transcriptional analysis. Bioinformatics [Epub ahead of print]. [abstract]