Cells tracked through four points in time to map activation at molecular level
In a first-of-its-kind experiment, researchers have identified links between 127 genes and immune diseases, providing newfound insights into the sequence and timing of gene activity during the activation of T cells, a key process in regulating the body’s immune response.
The study, led by researchers from Open Targets, the Wellcome Sanger Institute and GSK, is published today (26 May 2022) in Nature Genetics. The result of a 3 year research effort, it is the first to show that numerous disease-linked variants are active during different stages of T cell activation, providing key information to guide the development of new therapies for immune diseases such as rheumatoid arthritis, type-1 diabetes and Crohn’s disease.
T cells are a group of white blood cells that help to manage the human immune system. When T cells malfunction, they can cause severe immune deficiencies that leave patients at high risk of infection, as well as more common autoimmune diseases such as type 1 diabetes and rheumatoid arthritis, where the body mounts an attack against its own cells.
T cell activation is the first step in the immune system’s response to infection. Mapping this activation cycle at a molecular level is crucial to understanding where it can go wrong, and at which points therapeutic interventions can influence the process. But to date, only limited snapshots of the activation sequence have been mapped, such as looking at cells at a single time point. These snapshot techniques have also been unable to capture the diversity of cell subtypes present at each stage. Previous research from Open Targets showed that certain variations in DNA associated with immune diseases are linked to the T cells’ activation process1.
In this new study, researchers at the Wellcome Sanger Institute and GSK profiled over 650,000 individual cells using single-cell RNA sequencing technology to map the timing of gene activity for each cell subtype in the T cell activation process. They identified genes regulated by variations in DNA that were switched on or off in each cell, from their resting state through three different time points during T cell activation.
The team identified over 6,400 genes involved in the activation process. By comparing their data with known genetic variants for 13 immune diseases, they were able to find 127 genes associated with those diseases, some of which only manifested at specific time points that have not been previously studied.
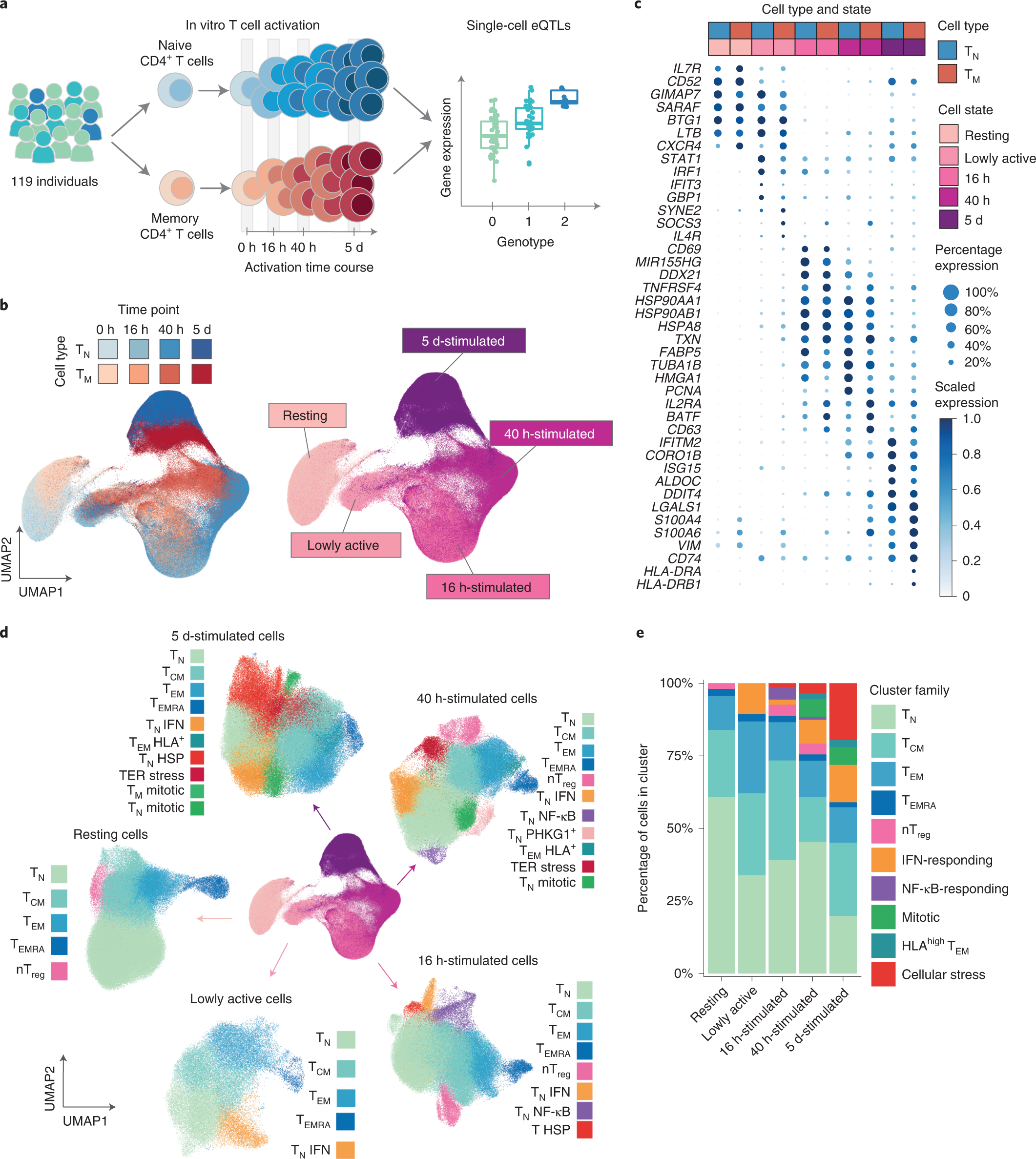
A single-cell transcriptional map of CD4+ T cell activation
a, Schematic of the study design. b, UMAP embedding of scRNA-seq data for unstimulated CD4+ T cells and at three time points after activation. Colors represent cell types (blue, naive T cell (TN); red, memory T cell (TM)), and shades of colors indicate time points (lighter shades for early time points and darker shades for late time points). Right panel represents the five broad cell states. c, Dot plot of highly variable gene expression throughout T cell activation. Shades of blue represent average expression in each cell population, and dot sizes represent the proportion of cells expressing the gene. d, Separate UMAP embeddings for the five broad cell states. Colors represent cell populations derived from unsupervised clustering. e, Proportion of different cluster groups present at each time point. Cell populations defined from clustering were classified into one of ten families, represented in different colors. ER, endoplasmic reticulum; HLA, human leukocyte antigen; HSP, heat shock protein; NF-κB, nuclear factor κB; nTreg, natural (i.e. thymus-derived) regulatory T cells.
Dr Blagoje Soskic, a first author of the study from the Wellcome Sanger Institute, said: “By profiling multiple time points during T cell activation, our study emphasises that genetic regulation can be specific for a particular cell state. Crucially, we were able to associate DNA variants with changes in the activity of specific genes, in particular cell types, at various times in the activation process. This unprecedented granularity is key to better understanding T cell activation, providing more in-depth data with which to pursue new treatments for immune disorders.”
John Lepore, SVP and Head of Research at GSK, said: “This study is already informing our early discovery portfolio by providing a novel, rich data set we are actively using to select genetically-informed drug targets for further validation experiments. As a founding partner of Open Targets, GSK is pleased that studies like this one can help further our shared understanding of the intersection of human genetics and immunology so we can get ahead of disease together.”
While this study focused on genes involved in 13 immune diseases, the approach and data generated can be applied to identifying genes involved in other disorders. The authors hope that this will shed light on how diseases are related or underpinned by similar biological processes, and highlight genetic risk factors. Ultimately, this work informs drug discovery to create new and better therapies for immune-mediated conditions.
Dr Gosia Trynka, the senior author of the study from the Wellcome Sanger Institute and Experimental Science Director of Open Targets, said: “T cells are particularly dynamic cells, so being able to capture their activation process in such detail is an important achievement. Our data will provide crucial insight into key immune system mechanisms, helping us to understand which genes are affected, what might be causing disease, and what factors might be putting patients at risk. In particular, the ability to link genetic variants strongly tied to immune diseases with changes in the activity of genes during this activation is a crucial first step to developing new treatments.”
Source – Wellcome Sanger Institute
Soskic B, Cano-Gamez E, Smyth DJ, Ambridge K, Ke Z, Matte JC, Bossini-Castillo L, Kaplanis J, Ramirez-Navarro L, Lorenc A, Nakic N, Esparza-Gordillo J, Rowan W, Wille D, Tough DF, Bronson PG, Trynka G. (2022) Immune disease risk variants regulate gene expression dynamics during CD4+ T cell activation. Nat Genet [Epub ahead of print]. [article]