Deconvolution algorithms rely on single-cell RNA-sequencing (scRNA-seq) data applied onto bulk RNA-sequencing (bulk RNA-seq) to extract information on the cell-types composition and proportions comprising a certain tissue. Adipose tissues’ cellular composition exhibits enormous plasticity in response to weight changes and high variance at different anatomical locations (depots). However, adipocytes – the functionally unique cell type of adipose tissue, are not amenable to scRNA-seq, a challenge recently met by applying single-nucleus RNA-sequencing (snRNA-seq).
Researchers at the Ben-Gurion University of the Negev aimed to develop a deconvolution method to estimate the cellular composition of human visceral and subcutaneous adipose tissues (hVAT and hSAT, respectively) using snRNA-seq to assess the true cell-type proportions. To correlate deconvolution-estimated cell-type proportions to true (snRNA-seq -derived) proportions, the researchers analyzed seven hVAT and 5 hSAT samples by both bulk RNA-seq and snRNA-seq. snRNA-seq uncovered 15 distinct cell types in hVAT and 13 in hSAT. Deconvolution tools – SCDC, MuSiC, and Scaden exhibited low performance in estimating cell-type proportions (median |R|= 0.12 for estimated vs. true correlations). Notably, estimation accuracy somewhat improved by decreasing the number of cell-types groups, which nevertheless remained low (|R|<0.42).
The researchers therefore developed sNuConv, a novel method that employs Scaden, a deep-learning tool, trained using snRNA-seq – based data corrected by i. snRNA-seq/bulk RNA-seq highly-correlated genes, ii. corrected estimated cell-type proportions based on individual cell-type regression models. Applying sNuConv on their bulk RNA-seq data resulted in cell-type proportion estimation accuracy with median R=0.93 (range:0.76–0.97) for hVAT, and median R=0.95 (range:0.92–0.98) for hSAT. The resulting model was depot-specific, reflecting depot-differences in gene expression patterns. Thus, they present sNuConv, a novel, AI-based, method to deduce the cellular landscape of hVAT and hSAT from bulk RNA-seq data, providing proof-of-concept for producing validated deconvolution algorithms for tissues un-amenable to single-cell RNA sequencing.
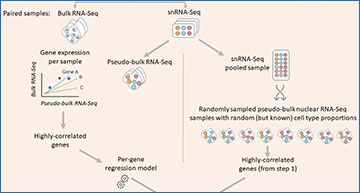
sNuConv workflow
sNuConv is a Scaden-based algorithm developed for estimating cell-type proportions from bulk RNA-seq data while training on snRNA-seq, rather than scRNA-seq, data. A-D denote the 4 stages of sNuConv, which include the conversion of snRNA-seq dataset into pseudo-bulk training set with per-gene correction (A.), generating a Scaden-based prediction model (B.), per cell-type regression model generation (C.) and correction (D.) to obtain the final sNuConv deconvolution output.
Sorek G, Haim Y, Chalifa-Caspi V, Lazarescu O, Ziv M, Hagemann T, Nankam PAN, Blüher M, Liberty IF, Dukhno O, Kukeev I, Yeger-Lotem E, Rudich A, Levin L. (2023) sNucConv: A bulk RNA-seq deconvolution method trained on single-nucleus RNA-seq data to estimate cell-type composition of human subcutaneous and visceral adipose tissues. bioRXiv [online preprint]. [article]