Researchers at Karolinska Institutet, Sweden have uncovered the molecular logic underpinning the assembly of spinal circuits that control the speed of locomotion in adult zebrafish. The study has recently been published in Nature Neuroscience.
What does the study show?
A fundamental hallmark of motor actions is the flexibility of their timing, speed and strength that is central to rapid adaptation to the ever-changing world around us. This is particularly apparent during locomotion, a behavior that involves full-body coordination characterized by sudden changes in speed and strength.
“In this study, we used single-cell RNA sequencing in adult zebrafish to link the molecular diversity of motoneurons and interneurons with their modular circuit organization that is responsible for changes in locomotor speed” says Abdel El Manira, Professor at the Department of Neuroscience at Karolinska Institutet, and corresponding author of the article.
“We show that each neuronal population comprises three specific subtypes defined by key molecular features and correspond to neurons underlying locomotion at slow, intermediate and fast speeds”.
Furthermore, the researchers’ analysis reveals molecular signatures that define each of the three circuit speed modules. The study uncovers the molecular underpinnings for neuronal diversity and how they relate to the function of locomotor circuits in adult zebrafish.
Molecular characterization of MN diversity
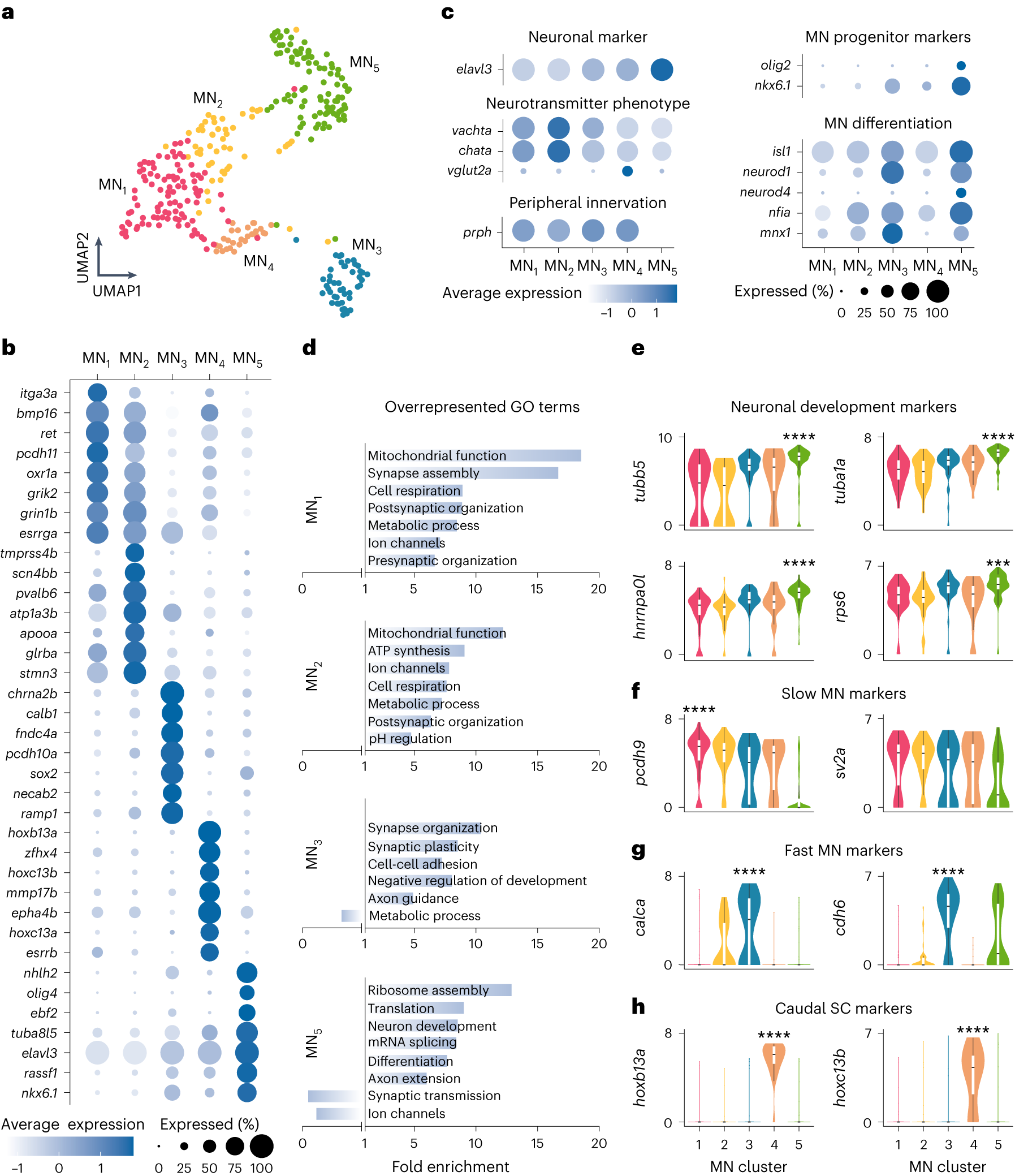
a, MN single-cell transcriptomes visualized using UMAP of five color-coded, molecularly defined clusters (n = 316 cells; MN1, n = 98; MN2, n = 51; MN3, n = 47; MN4, n = 27; MN5, n = 93). b, Examples of differentially expressed genes in each cluster. The size of the circle reflects the proportion (%) of cells expressing the gene, and the color intensity reflects its average expression level within that cluster. c, Normalized expression levels for motoneuronal marker genes. d, GO analysis of differentially expressed genes in each cluster. e, Log-normalized gene expression levels for neuronal development gene markers (differential gene expression analysis, nonparametric Wilcoxon rank sum test with Bonferroni adjusted P value, ***P = 0.0003, ****P < 0.0001). f, Log-normalized expression levels of known gene markers of slow MNs in zebrafish (pcdh9) or mice (sv2a) (differential gene expression analysis, nonparametric Wilcoxon rank sum test with Bonferroni adjusted P value, ****P < 0.0001). g, Log-normalized expression levels of known gene markers of fast MNs in mice (differential gene expression analysis, nonparametric Wilcoxon rank sum test with Bonferroni adjusted P value, ****P < 0.0001). h, Log-normalized expression levels for caudal spinal cord (SC) gene markers (differential gene expression analysis, nonparametric Wilcoxon rank sum test with Bonferroni adjusted P value, ****P < 0.0001). In e–h, boxes are bound by 25th and 75th percentiles, center line indicates the median and whiskers extend from minimum to maximum.
Why are these results important?
Overall, by characterizing how the molecular diversity of motoneurons and interneurons relates to their function, connectivity and behavior, the study provides important insights not only into the molecular mechanisms for neuronal and circuit diversity for locomotor flexibility but also for charting circuits for motor actions in general.
“While the categorization of muscle units into slow, intermediate, and fast types is universal to all vertebrates, the molecular underpinnings of motoneuron diversity and their premotor circuits have remained unclear. Our study fills this critical gap in our knowledge and represents a true advance in the field, says Irene Pallucchi, affiliated researcher and co-author of the article.
Moreover, the conceptual advance provided by the study is of broad interest to researchers in the motor control field and brain circuit organization in general.
How did you perform the study?
“We used single-cell RNA sequencing, electrophysiology, anatomical, and behavioral analysis in adult zebrafish, which provide both experimental and genetic accessibility”, explains Maria Bertuzzi, research engineer in Abdel El Manira group and co-author of the article.
This allowed to reveal the molecular and functional features that define motoneuron and interneuron subtypes, as well as their modular circuit organization responsible for controlling locomotion speed.
Gearshift mechanisms for speed change
The molecularly defined three-speed circuit module organization uncovered in the study acts as gearshift mechanisms for speed change and can also allow for rapid changes in direction. However, it is not known how the different speed circuit modules are driven from the brainstem.
“Our future studies aim to test the hypothesis that the commands from the brain are channeled to the spinal locomotor network through distinct command streams, each of which preferentially controls one of the speed circuit modules”, says Abdel El Manira. “Such an organization would ensure a high degree of flexibility and maneuverability of locomotor movements in a task- and context-dependent manner.”
The study was funded by the Swedish Research Council, Knut and Alice Wallenberg Foundation, Swedish Brain Foundation
Source – Karolinska Institutet
Pallucchi I, Bertuzzi M, Madrid D, Fontanel P, Higashijima SI, El Manira A. (2023) Molecular blueprints for spinal circuit modules controlling locomotor speed in zebrafish. Nat Neurosci [Epub ahead of print]. [article]