Single-cell RNA-sequencing (scRNA-seq) offers functional insight into complex biology, allowing for the interrogation of cellular populations and gene expression programs at single-cell resolution. Researchers at the University of Toronto have developed scPipeline, a single-cell data analysis toolbox that builds on existing methods and offers modular workflows for multi-level cellular annotation and user-friendly analysis reports. Advances to scRNA-seq annotation include: (i) co-dependency index (CDI)-based differential expression, (ii) cluster resolution optimization using a marker-specificity criterion, (iii) marker-based cell-type annotation with Miko scoring, and (iv) gene program discovery using scale-free shared nearest neighbor network (SSN) analysis. Both unsupervised and supervised procedures were validated using a diverse collection of scRNA-seq datasets and illustrative examples of cellular transcriptomic annotation of developmental and immunological scRNA-seq atlases are provided herein. Overall, scPipeline offers a flexible computational framework for in-depth scRNA-seq analysis.
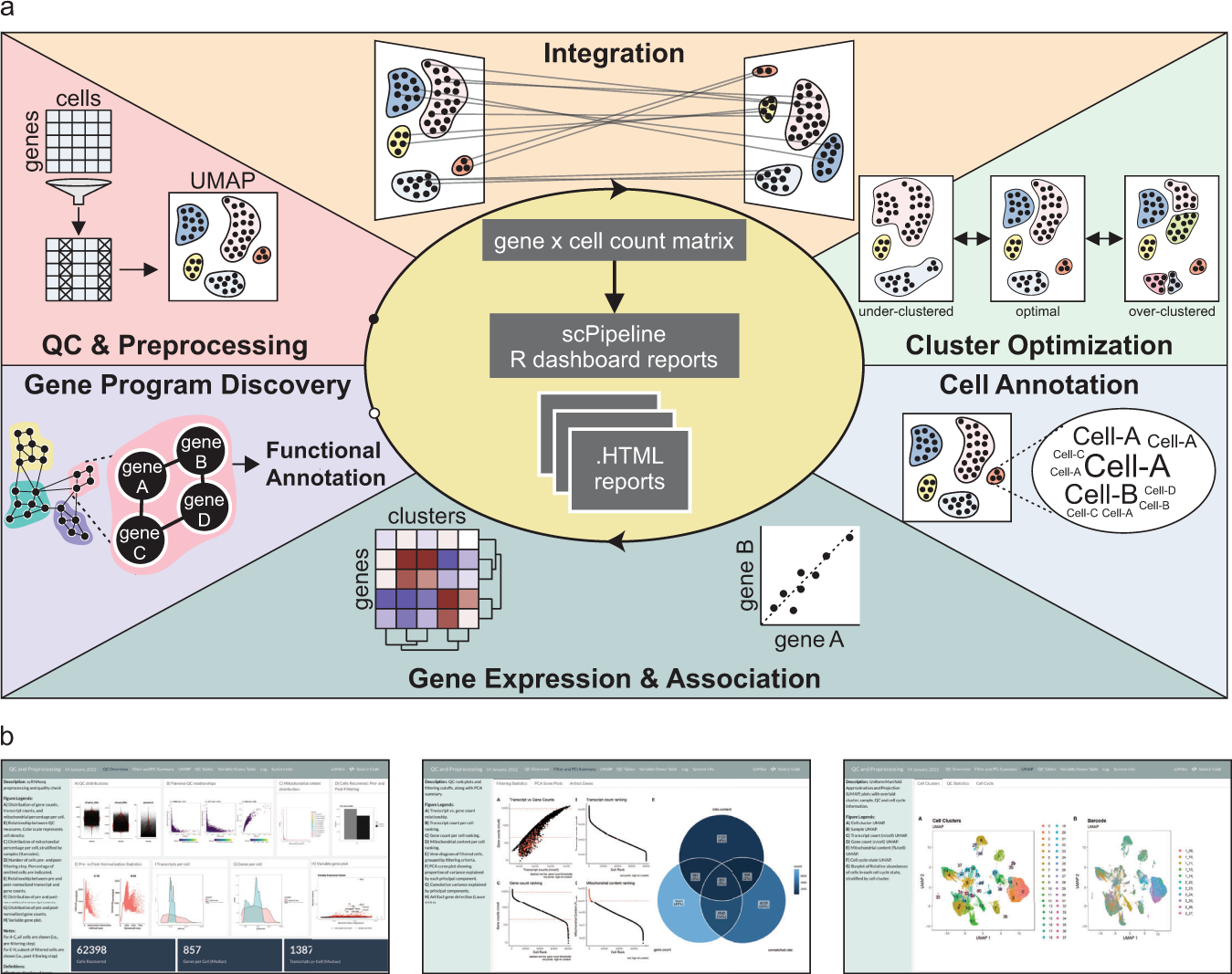
Schematic of scPipeline analysis modules
a scPipeline is a modular collection of Rmarkdown scripts that generate reports for scRNA-seq analyses. The modular framework permits flexible usage and facilitates i) QC & preprocessing, ii) integration, iii) cluster optimization, iv) cell annotation, v) gene expression and association analyses, and vi) gene program discovery. Each standalone HTML report provides a comprehensive analysis summary that can be seamlessly shared without any dependencies. Alternatively, online repositories (e.g., GitHub) can be used to host HTML reports for public dissemination. b Representative snapshots of scPipeline reports generated using the QC and preprocessing (left), cell annotation (middle), and gene expression and association (right) modules.
Availability – The scMiko R package (https://github.com/NMikolajewicz/scMiko) and scPipeline scripts (https://github.com/NMikolajewicz/scPipeline) are available on GitHub.
Mikolajewicz N, Gacesa R, Aguilera-Uribe M, Brown KR, Moffat J, Han H. (2022) Multi-level cellular and functional annotation of single-cell transcriptomes using scPipeline. Comm Biol 5: 1142. [article]