Environmental enteropathy (EE), or gut inflammation due to environmental factors, is thought to be a major contributor to growth stunting, malnutrition, and reduced oral vaccine responses in millions of children and adults in low-resource settings. The condition is often regarded as secondary to other issues such as frequent intestinal infections and poor access to sanitation infrastructure and clean water. While it is appreciated that gut inflammation can occur due to exposure to contaminated food or water, causing changes that make it difficult for children to absorb nutrients from food properly, exactly what those changes are at a cellular and molecular level remains poorly understood. In a recent paper in Science Translational Medicine, researchers at the Ragon Institute of MGH, MIT and Harvard; University Teaching Hospital Lusaka, Zambia; Queen Mary University of London; the University of California, San Francisco; and SUNY Downstate Health Sciences University used single-cell profiling to define the impact on the intestine of exposure to adverse environmental conditions in a disadvantaged community in Lusaka, Zambia. Their work provides a roadmap for future EE intervention studies.
The researchers examined 33 small intestinal biopsies from 11 adults with EE in Lusaka, Zambia (eight HIV-negative and three HIV-positive), six adults without EE in Boston, USA, and two adults in Durban, South Africa. They also incorporated published data on three additional individuals from the Durban site. They used single-cell RNA-sequencing, a technique that profiles the RNA levels of individual cells to better understand their characteristics and functions, to help uncover differences seen in the gut of individuals with EE.
“Environmental enteropathy is in many ways a perfect application for single-cell RNA-sequencing,” says Conner Kummerlowe, PhD, a postdoctoral researcher at the Ragon Institute and lead author of the paper, adding that the condition has been systematically understudied. “To be able to just really unbiasedly go in and see what’s happening in this condition that’s poorly understood is really powerful.”
Single-cell RNA-sequencing of the upper small intestine with and without EE
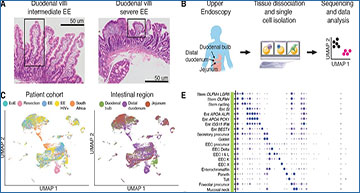
A, H&E imaging of the small intestine with intermediate and severe EE. Intestinal villi with normal morphology (left image) and with severe blunting (right image) are highlighted in boxes. B, Experimental workflow: small intestinal biopsies from the duodenal bulb, distal duodenum, and jejunum were obtained via endoscopy or tissue resection, dissociated into cells, loaded onto a Seq-Well array, processed for single-cell sequencing, and analyzed. C, UMAP visualization by patient cohort and intestinal region for all 26,556 high quality cells from 38 samples and 22 patients. D, Fractional abundances of major cell types amongst all single cells analyzed per sample. E, Expression of marker genes for final cell subsets. Dot size represents the fraction of a cell subset (rows) expressing a given gene (columns). Dot hue represents the scaled average expression by gene column. For clarity, dots for genes expressed in 5% or less of cells within a given subset are not shown.
Previous research that analyzed EE biopsies histologically showed that in the small intestines of those with EE, the villi — structures that absorb nutrients from food — are shorter. It was also found that the gut’s barrier functions are responding in a pattern that resembles an attempt to keep harmful bacteria out, but which has the side effect of preventing nutrient absorption. In the new study, the researchers found evidence that this may be due to gut intestinal epithelial cells differentiating into surface mucosal cells – mucus-secreting cells normally found in the stomach help to protect against acid-associated damage. Furthermore, the team observed that these cells express antimicrobial genes including DUOX2, which had previously been described as a key marker of EE.
Surface mucosal cells are normally not found in the small intestine. One theory the researchers have is that mucosal cells may build up on the tips of the villi during wound-healing responses to infections from commonly occurring bacteria such as Helicobacter pylori.
“The real power of methods like single-cell RNA-sequencing is that they take a global view of disease processes at the level at which biology operates – the level of the cell. Here, this let us identify unexpected cellular behaviors that we may be able to target to improve nutrient absorption,” says Alex K. Shalek, PhD, co-senior author of the paper and a Member of the Ragon Institute. Shalek is also a Core Member of the Institute for Medical Engineering and Science (IMES), an Associate Professor of Chemistry, and an Extramural Member of the Koch Institute at MIT, as well as an Institute Member of and member of the Broad Institute of Harvard and MIT.
By comparing EE samples with control cohorts, they also identified dysregulated WNT and MAPK cell signaling pathways and increased expression of proinflammatory genes in a subset of memory T cells.
“Comparing these cell profiles allowed us to identify perturbations in absorptive cell replication and differentiation alongside specific changes in tissue resident memory T cells. These changes align with known defects in the function of the intestine in individuals with EE,” says Paul Kelly, MD, Professor of Tropical Gastroenterology at Queen Mary University of London and co-senior author of the study.
The researchers also studied the potential impact of HIV infection and antiretroviral treatment on the pathology of EE. They found that samples from HIV+ individuals had higher EE severity and that there were telltale signs of HIV infection expressed in the duodenal bulb, the part of the small intestine located closest to the stomach.
The work identifies correlates of EE that may be cellular and molecular targets for intervention in the future.
“Our findings have identified multiple possible therapeutic and prophylactic approaches, as well as suggested possible mechanisms to explain the efficacy of prior interventions that showed benefit,” says Thomas Wallach, MD, physician-scientist and pediatric gastroenterologist at SUNY Downstate Health Sciences University and co-senior author, who completed the research as a pediatric gastroenterology fellow at the University of California, San Francisco Medical Center. He further noted “our observation of signals linked with immune tolerance may also shed light onto mechanisms underlying the hygiene hypothesis,” a possible explanation of the relative rarity of autoimmune and allergic diseases in low-middle income countries.
Source – SUNY Downstate Health Sciences University
Kummerlowe C, Mwakamui S, Hughes TK, Mulugeta N, Mudenda V, Besa E, Zyambo K, Shay JES, Fleming I, Vukovic M, Doran BA, Aicher TP, Wadsworth MH 2nd, Bramante JT, Uchida AM, Fardoos R, Asowata OE, Herbert N, Yilmaz ÖH, Kløverpris HN, Garber JJ, Ordovas-Montañes J, Gartner ZJ, Wallach T, Shalek AK, Kelly P. (2022) Single-cell profiling of environmental enteropathy reveals signatures of epithelial remodeling and immune activation. Sci Transl Med 14(660):eabi8633. [abstract]