Sometimes, in the quest for new data, scientists forget that existing datasets can be treasure troves for new discoveries. One such undermined scientific gem is transcriptomic data from the human brain, both with and without gliomas.
“A lot of bulk RNA-seq data is already publicly available,” explained Sonali Arora, a computational biologist in the Holland lab in the Human Biology Division at Fred Hutchinson Cancer Center. “By combining these publicly available datasets, we can really harness their collective power to understand and explore cancer without re-sequencing tumors.”
And that’s exactly what she did. In a new paper published in Scientific Reports, Arora and colleagues describe a reference landscape of the human brain compiled from five publicly available large-scale datasets. The landscape, called the Brain-UMAP, is freely accessible on the open-source website Oncoscape as an interactive online tool. Oncoscape is a web-based data exploration and visualization tool developed by the Seattle Translational Tumor Research, a research initiative drawing on the expertise of oncologists and cancer researchers at Fred Hutch, UW Medicine, and Seattle Children’s.
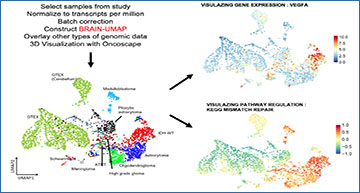
To create this reference landscape of the brain, Arora and colleagues identified five existing large-scale glioma datasets. These datasets included three of adult gliomas, one of pediatric gliomas, and one representing healthy adult brains. All five datasets had transcriptomic data from RNA-seq, and three had additional genomic data from whole-genome sequencing. The authors applied Uniform Manifold Approximation and Projection (UMAP) to the dataset to assemble a reference Brain-UMAP. To allow for 3-dimensional visualization of the data, Matt Jensen, the senior data visualization engineer for Oncoscape and a co-author on the new paper, incorporated the five datasets into Oncoscape. As an interactive tool, Oncoscape allows users to visualize and manipulate the Brain-UMAP.
Researchers used five publicly available large-scale datasets to develop the Brain-UMAP, a reference landscape of healthy and neoplastic brain. The Brain-UMAP, freely accessible on the open-source website Oncoscape, allows researchers to explore and visualize both gene- and pathway-level differences between healthy brains and brains with gliomas. Image provided by Sonali Arora
This reference landscape allows researchers to explore and visualize both gene- and pathway-level differences between healthy brains and brains with glioblastomas. The authors found that the glioma dataset and the dataset from healthy brains separated into distinct clusters. The pediatric glioma dataset fell in between two adult clusters. Furthermore, applying the Brain-UMAP to the healthy brain samples indicated two distinct clusters of brain regions: the supratentorial regions and the cerebellum. In addition, the authors used the three datasets with genomic and transcriptomic data to explore copy number, mutations and gene fusions at a single-gene level.
These findings are just the tip of the iceberg when it comes to the trove of information that can be mined from this dataset.
“Researchers studying not only brain cancer such as glioblastoma, but also a vast majority of pediatric studies, are often interested in a single gene or a single pathway,” said Arora. “Our reference landscape is freely accessible in Oncoscape, and the research community can interact with the data. They can readily interact with our landscape and explore how their gene or pathway of interest behaves.”
Cancer researchers are already taking advantage of the Brain-UMAP generated in this study. So far, studies that utilize the Brain-UMAP include two papers from the Paddison lab at Fred Hutch (a paper by Mitchell et al. describing a gene for potential therapeutic target for glioblastomas; a functional-genomic analysis comparing and contrasting adult and pediatric glioblastomas by Hoellerbauer et al.), as well as an ongoing project led by Nicholas Nuechterlein, a PhD student in the Holland lab and a co-author on the current paper.
Although this study focused on brain tumors, the authors hope to leverage this tool to understand other cancers, and even diseases beyond cancer.
“We are interested in creating similar reference landscapes for other cancer types and disease models, and see if our research can benefit researchers worldwide,” explained Arora.
Open-source tools like Oncoscape allow researchers to take advantage of existing large-scale datasets to improve patient diagnoses and prognoses, as well as gain a deeper understanding of the underlying biology of cancer.
“As researchers, we can get so focused on comparing like with like that we lose sight of the proverbial forest for focusing too much on the leaves of a single tree,” the authors wrote in the paper. “By integrating multiple datasets while correcting for batch effects, such as with the Brain-UMAP presented here, we can harness the power of multiple datasets.”
Source – Fred Hutch Cancer Center
Availability – Brain-UMAP is freely accessible on the open-source website Oncoscape as an interactive online tool.
Arora S, Szulzewsky F, Jensen M, Nuechterlein N, Pattwell SS, Holland EC. (2023) Visualizing genomic characteristics across an RNA-Seq based reference landscape of normal and neoplastic brain. Sci Rep 13(1):4228. [article]