Clonal bacterial populations rely on transcriptional variation across individual cells to produce specialized states that increase fitness. Understanding all cell states requires studying isogenic bacterial populations at the single-cell level. A team led by researchers at IFF Health and Biosciences have developed probe-based bacterial sequencing (ProBac-seq), a method that uses libraries of DNA probes and an existing commercial microfluidic platform to conduct bacterial single-cell RNA sequencing.
The research team sequenced the transcriptome of thousands of individual bacterial cells per experiment, detecting several hundred transcripts per cell on average. Applied to Bacillus subtilis and Escherichia coli, ProBac-seq correctly identifies known cell states and uncovers previously unreported transcriptional heterogeneity. In the context of bacterial pathogenesis, application of the approach to Clostridium perfringens reveals heterogeneous expression of toxin by a subpopulation that can be controlled by acetate, a short-chain fatty acid highly prevalent in the gut.
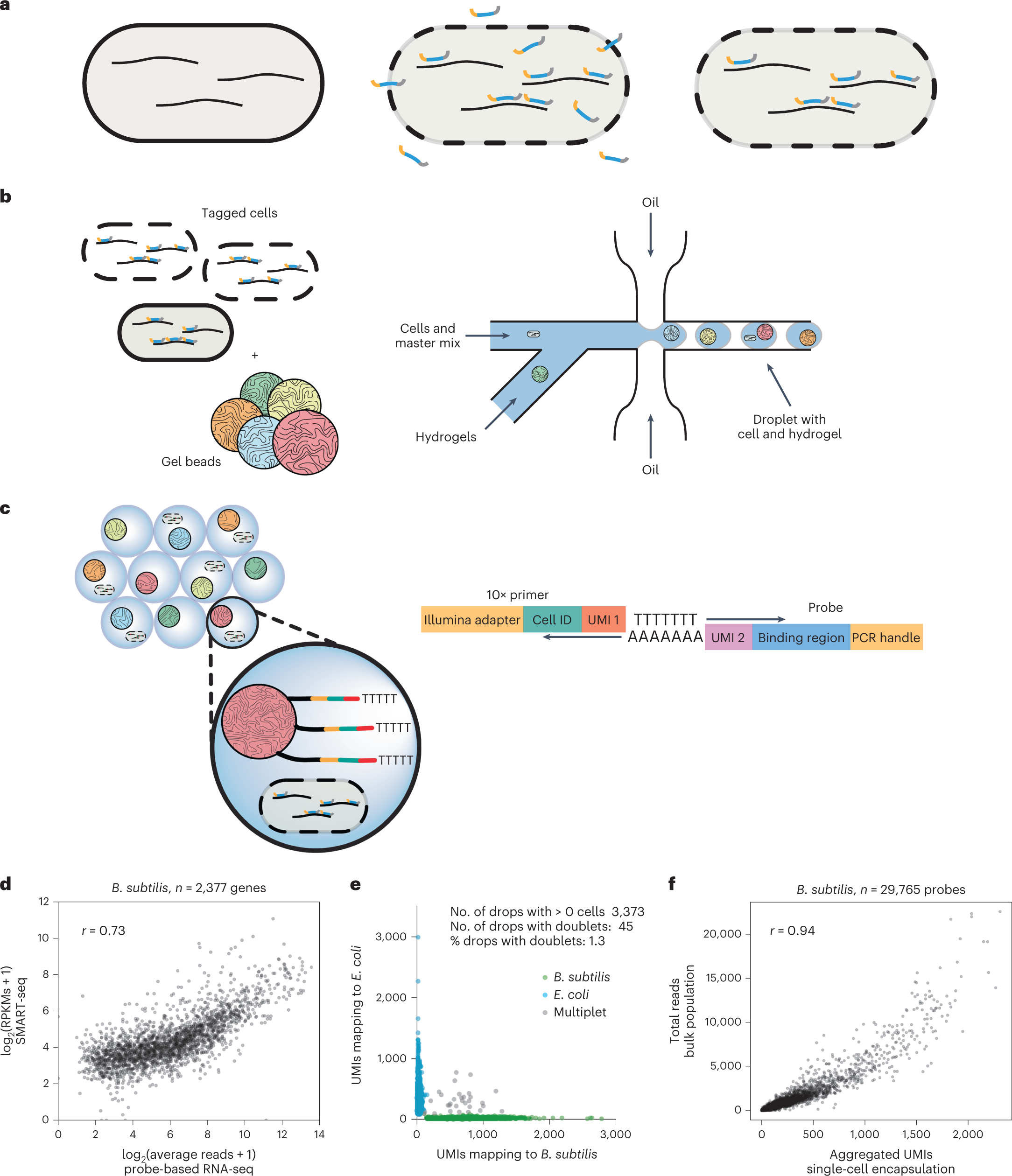
Microfluidic probe-based scRNA-seq method and validation
a, Cells were fixed and permeabilized to allow the penetration of thousands of unique, genome-specific oligonucleotide probes. Hybridized probes retrofitted transcripts with a poly-A tail and UMI, whereas unhybridized probes were washed away. b, Permeabilized cells with hybridized probes were flowed through a commercial microfluidic device that encapsulates single cells into droplets containing barcoded primers with poly-A capture sequence conjugated to a hydrogel microsphere and PCR reagents. c, Final droplets contain one or fewer cells and one hydrogel with a unique cell barcode. Barcoded cDNA was generated from the mRNA:probe hybridized complex via in-droplet PCR. Droplets were then broken and the pooled cDNA amplified further before sequencing. Single-cell transcriptomes were resolved, clustered and visualized. d, Transcriptome quantification by hybridization of a probe library followed by PCR correlates (Pearson’s correlation coefficient, r = 0.73) to traditional, bulk RNA-seq method (SMART-seq stranded kit, Takara) involving random priming of hexamers followed by reverse transcription (RT) and incorporation of template switching oligo. e, Species mixture (‘barnyard’) plot demonstrates that single cells of different bacterial species can be resolved by barcode after microfluidic encapsulation. f, Aggregated probe-based signal from thousands of single cells is well correlated (Pearson’s correlation coefficient, r = 0.94) to the average probe-based signal obtained from the bulk population (pre-encapsulation). RPKM, reads per kilobase of exon per million reads mapped.
Overall, ProBac-seq can be used to uncover heterogeneity in isogenic microbial populations and identify perturbations that affect pathogenicity.
McNulty R, Sritharan D, Pahng SH, Meisch JP, Liu S, Brennan MA, Saxer G, Hormoz S, Rosenthal AZ. (2023) Probe-based bacterial single-cell RNA sequencing predicts toxin regulation. Nat Microbiol [Epub ahead of print]. [article].