Human development starts from the fertilization of a fully matured oocyte. Oocyte maturation, fertilization, and early embryo development before zygotic activation is characterized by the absence of new transcription from the genome. Thus, the process of oocyte-to-embryo transition before zygotic genome activation is largely controlled by post-transcriptional regulation of maternal mRNA stored in the fully grown oocyte. However, how maternal mRNA is post-transcriptionally regulated has remained elusive.
Researchers led by Prof. LU Falong at the Institute of Genetics and Developmental Biology (IGDB) of the Chinese Academy of Sciences (CAS), together with collaborators, have utilized poly(A)-inclusive RNA isoform sequencing (PAIso-seq) and PAIso-seq2 methods to comprehensively profile the poly(A)-tail-inclusive full-length transcriptome in human oocytes and embryos.
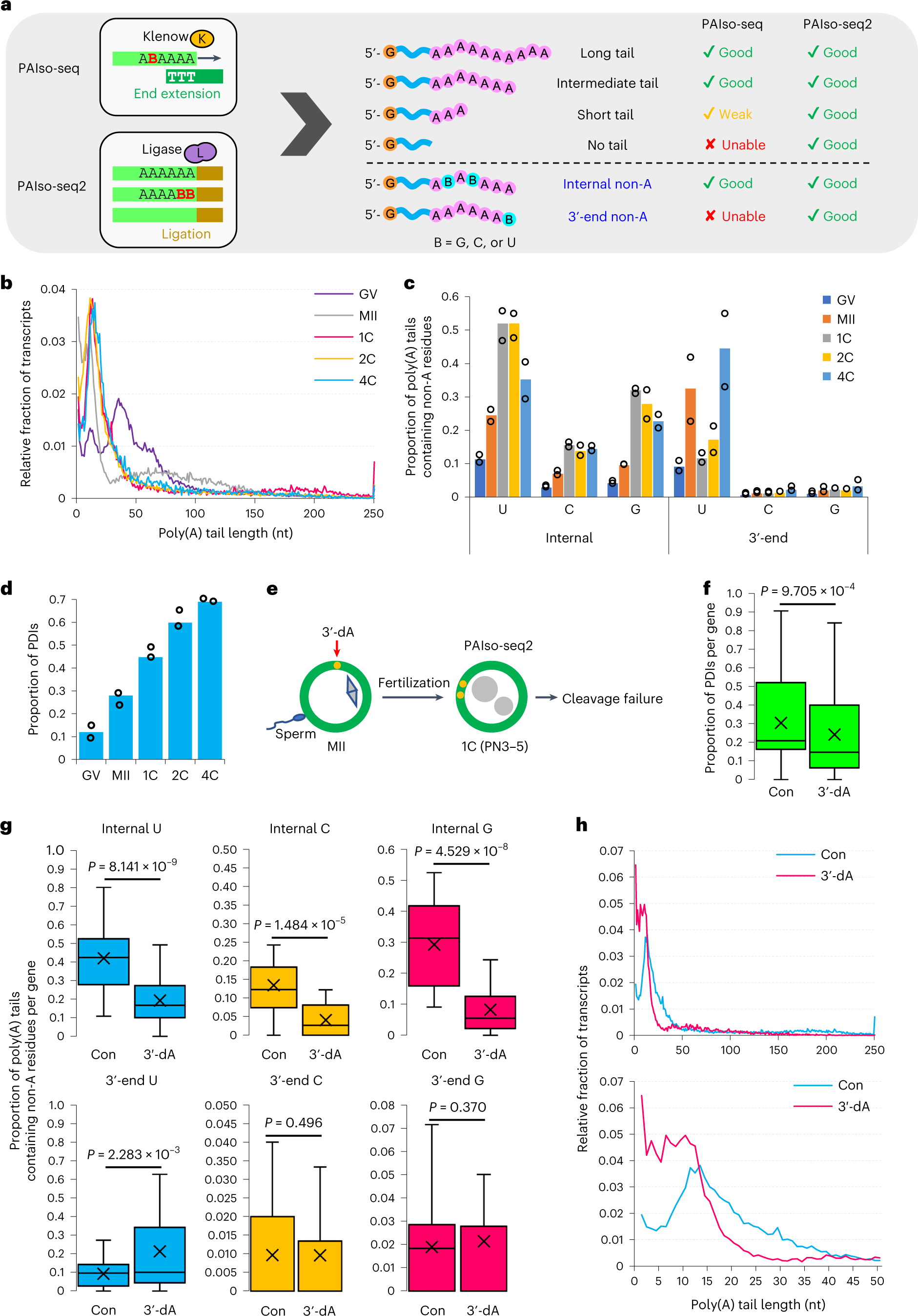
Human oocytes and early embryos analyzed by PAIso-seq2
a, Diagram showing the principle and capability of capturing different types of transcripts for PAIso-seq and PAIso-seq2. b, Global distribution of poly(A) tail lengths of mRNAs measured by PAIso-seq2. Median poly(A) tail length (nt): GV, 40; MII, 25; 1C, 21; 2C, 17; 4C, 21. c, Frequency of non-A residues separated by internal and 3′-end positions in oocytes and early embryos, measured by PAIso-seq2 (two replicates each). d, The PDI level of mRNAs in oocytes and early embryos, measured by PAIso-seq2 (two replicates each). e, Illustration of 3′-dA treatment experiments in 1C embryos for PAIso-seq2 analysis. f, Box plots showing the PDI level of individual genes (29 genes with ≥20 reads in both samples) in control (Con) and 3′-dA-treated 1C embryos. g, Box plot of frequency of non-A residues of individual genes (29 genes with ≥20 reads in both samples) in control and 3′-dA-treated 1C embryos. h, Global distribution of poly(A) tail lengths of mRNAs in control and 3′-dA-treated 1C embryos. The bottom histogram is zoomed in view of the upper histogram between 1–50 nt. The data here were measured by PAIso-seq2. Error bars indicate the s.e.m. The P values were calculated by one-tailed Student’s t-test. Genes with ≥20 reads were included for gene-level analysis. For all box plots, the ‘×’ indicates the mean value, the horizontal bars show the median value, and the top and bottom of the box represent the value of 25th and 75th percentiles, respectively.
The researchers found extensive poly(A)-tail-mediated maternal mRNA remodeling during the human oocyte-to-embryo transition. Unexpectedly, a large proportion of the maternal mRNA undergo partial degradation into their 3’-untranslated regions, which can then be modified by incorporation of uracil residues at their 3’-ends.
These transcripts do not undergo degradation and can be re-polyadenylated after fertilization, contributing to the production of a large number of new types of mRNA transcripts called polyadenylated degradation intermediates.
The researchers then revealed several regulators of this extensive maternal mRNA remodeling process, including BTG4 in regulating global deadenylation during oocyte maturation, TUT4/7 in contributing to the incorporation of U residues, and TENT4A/B in incorporation of G residues during the re-polyadenylation after fertilization that likely stabilized the re-polyadenylated mRNA transcripts.
What are the roles of these incorporated uracil residues in the poly(A) tails and what are the roles of the polyadenylated degradation intermediates? These are very interesting questions for the future.
Importantly, re-polyadenylation after fertilization is essential for human embryo development as evidenced by failed first embryo cleavage if the re-polyadenylation is blocked.
These findings provide rich insight into the poly(A)-tail-mediated post-transcriptional regulation of maternal mRNAs in regulating the human oocyte-to-embryo transition. This mechanism will offer a new angle for understanding the mechanism underlying female infertility in humans.
The poly(A)-inclusive full-length transcriptome will likely be a potentially useful tool in evaluating the developmental potential of oocytes, which could be valuable for infertility patients.
Source – Chinese Academy of Sciences
Liu Y, Zhao H, Shao F, Zhang Y, Nie H, Zhang J, Li C, Hou Z, Chen ZJ, Wang J, Zhou B, Wu K, Lu F. (2023) Remodeling of maternal mRNA through poly(A) tail orchestrates human oocyte-to-embryo transition. Nat Struct Mol Biol [Epub ahead of print]. [article]