Simple CUT&Tag adaptation gives information about genetic regulatory elements in formalin-fixed tumor samples
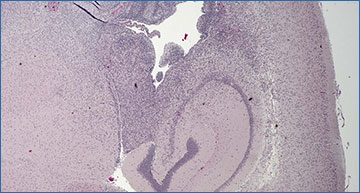
Formalin-fixed, paraffin-embedded (FFPE) tissue samples allow scientists to view tissue structure. Here, blue glioma cells invade normal brain tissue (in pink) from a brain cavity called a ventricle (the open area in the middle of the slide). The latest CUT&Tag adaptation gives scientists a new view: into gene regulation of FFPE-preserved tissue.
Precise, tailored cancer diagnosis and treatment, known as precision oncology, is gaining steam. But most methods able to look deeply at the molecular characteristics of a patient’s tumor rely on fresh or frozen cells — and it’s rare that surgically removed tumor tissue gets this treatment.
Tumor samples usually get soaked in formalin and then embedded in paraffin wax, ready for a pathologist to examine under a microscope. Unfortunately, it’s difficult to look for DNA mutations or changes in how genes are regulated in formalin-fixed, paraffin-embedded, or FFPE, tissue samples.
Now that may be changing. Fred Hutchinson Cancer Center scientists recently published a method in Nature Communications that adapts a technique called CUT&Tag to allow investigators to examine key gene regulatory elements in FFPEs.
“The data we get is so good that we can we can use it to identify the key genes that are involved [in tumors],” said Fred Hutch molecular biologist Steven Henikoff, PhD, who led development of the original CUT&Tag method and modified it for use on FFPEs.
Henikoff initially developed his new strategy, dubbed CUTAC, during the COVID-19 pandemic. CUTAC works around DNA packaging proteins to reveal regions that are important for regulating transcription, the process of turning information stored in DNA into RNA. Henikoff’s latest modifications enable its use on FFPEs.
Using CUTAC on mouse FFPEs, Henikoff and his collaborators were able to reveal previously unknown patterns of gene regulation in cancer. This included highlighting certain small, non-protein coding RNAs (called microRNAs) that may play a role in cancer and are difficult to detect using other methods.
The ability to mine molecular information from FFPEs “could allow us to do retrospective studies that link tumor biology with patient outcomes using samples already collected and stored,” said co-author and Fred Hutch brain cancer researcher Eric Holland, MD, PhD, who heads the Human Biology Division and holds the Endowed Chair in Cancer Biology.
The new method could help scientists explore biological processes beyond cancer, the researchers said. They are working with EpiCypher, Inc., the commercial provider of CUT&Tag that holds the licenses for CUT&Tag and adapted methods, to commercialize the newest method for use in FFPEs.
FFPEs: potential treasure trove of tumor molecular information
Many patients with cancer receive surgery or biopsies as part of their care. To preserve them for long-term storage and review by pathologists, it’s standard practice to soak these tissue samples in formalin and then embed them in paraffin. Scientists and physicians have been creating formalin-fixed, paraffin-embedded tissue samples, or FFPEs, since the late 19th century.
“Every pathology department in every hospital is full of these,” Holland said.
In the past decade or two, technological advances have made it possible for scientists to go far beyond a microscope-based look-see at tumor tissue. They can get detailed peeks inside individual cells, learning about gene sequences, which genes are turned on and off, and even how the DNA is packaged and bundled to regulate gene expression — all of which can have powerful influences on cancer development and progression.
But these technologies work on fresh or frozen cells — leaving the wealth of tumor molecular information stored in FFPEs nearly unreadable.
“But if you want a future in which we sequence everyone’s tumor and do something with that information, we can’t change everything. We need to start with what oncologists are using,” Holland said. “A group of us [at Fred Hutch]have been trying to get sequencing to work out of paraffin.”
Henikoff was interested in getting information that goes beyond the DNA sequence. The DNA packaging proteins that help it fit within a cell, and the molecular modifications that help regulate gene activation are called chromatin. Chromatin works like a computer program, helping orchestrate which genes are turned on and off.
CUT&Tag, for Cleavage Under Targets & Tagmentation, reveals chromatin patterns in DNA. Henikoff and his team developed the method to map chromatin in single cells. During the pandemic, he simplified the protocol to allow researchers to map more loosely packaged “accessible” chromatin areas at home. This adaptation was dubbed CUTAC, for Cleavage Under Targeted Accessible Chromatin.
Henikoff discovered that with a couple of simple changes to CUTAC, he could map regions of compacted chromatin and also where RNA-building enzymes (RNA polymerase II) are paused on DNA in FFPEs. These enzymes pause at regions that regulate transcription, the process of using a DNA sequence to build an RNA molecule. The pattern of active regulatory regions in a tissue can give important clues about the biological processes occurring — and whether they’ve changed in the tumor.
“It gives you the ultimate checkpoint for gene regulation. So if something goes wrong in cancer, it’s going to go wrong at the transcriptional level. It’s going to go wrong here,” Henikoff said. “We’re using RNA pol-II to tell us about regulatory elements and how they change in cancers.”
Turning disadvantages into advantages
Henikoff wasn’t the first to try CUT&Tag on FFPEs. Another group had already tried — and failed — with the original, in-lab protocol. Henikoff instead adapted his simpler at-home protocol, CUTAC.
“The problem is the formalin — it turns chromatin into concrete,” Henikoff said.
This is because formalin causes the amino acid lysine to form physical links to DNA; any lysine-bearing protein that binds DNA, whether to package it or help regulate a gene, is now firmly attached. This prevents scientists from sequencing this DNA. Dissolving these bonds using chemicals or enzymes further damages DNA.
There was another potential hurdle: Henikoff’s at-home CUT&Tag method produces small snippets of DNA, the short sections of “open” chromatin that lie between DNA packaging proteins.
“I thought, rather than fight it, why not take advantage?” Henikoff said.
He realized that his at-home CUT&Tag protocol was ideal for picking up the short “open” sections of DNA that lie between DNA packaging proteins and aren’t crosslinked by formalin.
Henikoff also discovered a gentle way to remove the paraffin that avoids DNA-damaging chemicals: heat. “Cooking” slides of FFPEs at about 85 degrees Celsius (approximately 185 degrees Fahrenheit) for a few hours melts away the paraffin and even helps reverse the bonds between DNA and proteins.
Revealing unknown cancer biology
Henikoff tested CUTAC on FFPEs of mouse brain and liver tumors as well as normal tissue. Holland’s team provided the brain tissue FFPEs. Ronald Paranal, MD, a hematology/oncology fellow in the lab of Fred Hutch co-author and liver and pancreatic cancer researcher Sita Kugel, PhD, compared liver tumors with normal liver tissue. The scientists found that CUTAC allowed them to distinguish different tumor types based on how different regulatory elements were up- or down-regulated. They were also able to distinguish between normal and tumor tissue.
CUTAC revealed more than the regulation of genes. While RNA is famous for being an intermediate step between genes and proteins, most of the RNA molecules our cells make don’t encode proteins. These non-coding RNAs, which can come in many sizes, still play important roles within cells.
Using CUTAC, Henikoff and Holland discovered that certain microRNAs, short non-protein coding RNAs, appear to play a role in cancer. One microRNA has a long evolutionary history and is shared among mice, humans, flies and even nematodes. It was already known to play a key role in glial cells, the supportive cells that help neurons work, but this study was the first to link the microRNA to brain cancer.
These microRNAs can’t be detected using standard RNA sequencing techniques, but CUTAC can infer their importance from the DNA regions that regulate their creation.
Expanding CUTAC’s applications
Given the ubiquity of FFPEs, the scientists are excited about the wide range of biological questions that CUTAC could be used to probe. Henikoff plans to explore ramped up transcription in cancer, as well as examine developmental processes using intestinal stem cells growing in the lab in 3D, which better approximates tissue structure than 2D plates.
And because CUTAC is simple and doesn’t require high-tech gadgetry, Henikoff is teaming up with Danielle Vermaak, PhD, former postdoctoral fellow from his lab to use the method to teach her high school students about gene regulation.
As a cancer biologist with an abiding interest in bringing precision oncology to the masses, Holland sees CUTAC as a step toward a future in which any patient’s tumor — no matter how it’s stored — can be analyzed for molecular clues to its critical genes and potential vulnerabilities. This information could be used to guide and tailor the patient’s cancer treatments.
Henikoff is also collaborating with Fred Hutch colleague Sanjay Srivastan, PhD, to expand CUTAC’s horizons, by using it on slides to detect spatial patterns of gene regulation in tissue samples.
“We have a method that we can use well beyond cancer and look at developmental processes and other diseases as well,” Henikoff said.
Source – Fred Hutchinson Cancer Center
Henikoff S, Henikoff JG, Ahmad K, Paranal RM, Janssens DH, Russell ZR, Szulzewsky F, Kugel S, Holland EC. (2023) Epigenomic analysis of formalin-fixed paraffin-embedded samples by CUT&Tag. Nat Commun 14(1):5930. [article]