Researchers at Karolinska Institutet have developed a molecular method able to detect whether or not bacteria respond to antibiotics within minutes. The findings are presented in the journal Nature Microbiology and the researchers now hope to develop a simple test for doctors to use.
“We are confident and hope that this can be one of many tools that doctors need to tackle antibiotic resistance, which is a serious and growing problem,” says principal investigator Vicent Pelechano, associate professor at the Department of Microbiology, Tumor and Cell Biology, Karolinska Institutet.
The method is called 5PSeq, is simple to use, and is based on sequencing the messenger RNA (mRNA) that the bacteria break down as they synthesise proteins. The measurements reveal how the bacteria are affected by different environmental factors, such as antibiotic treatments and other types of stress.
Rapid test for clinical use
The researchers tested the method on a total of 96 bacterial species from different phyla in complex clinical samples taken, for example, from faecal matter and the vagina, but also in compost samples. After only a matter of minutes, they were able to see whether or not the bacteria responded to antibiotic treatment; the effect was most salient after about half an hour.
3N Bio is a company that Dr Pelechano and some of his colleagues have started up in the KI Innovations incubator to develop the method and create a rapid molecular test for clinical use. They have now received financing from the Swedish Research Council to demonstrate proof of concept for such a test in collaboration with the Karolinska University Hospital.
“It’s crucial that doctors can quickly find the right antibiotics for seriously ill patients with bacterial infections to reduce the unnecessary use of antibiotics,” says Dr Pelechano. “Current methods of testing antibiotic resistance can take hours or even days, but often treatment needs to be given more promptly than that to avoid serious consequences for the patient. Because of this, a broad-spectrum antibiotic is often prescribed, which increases the risk of resistance.”
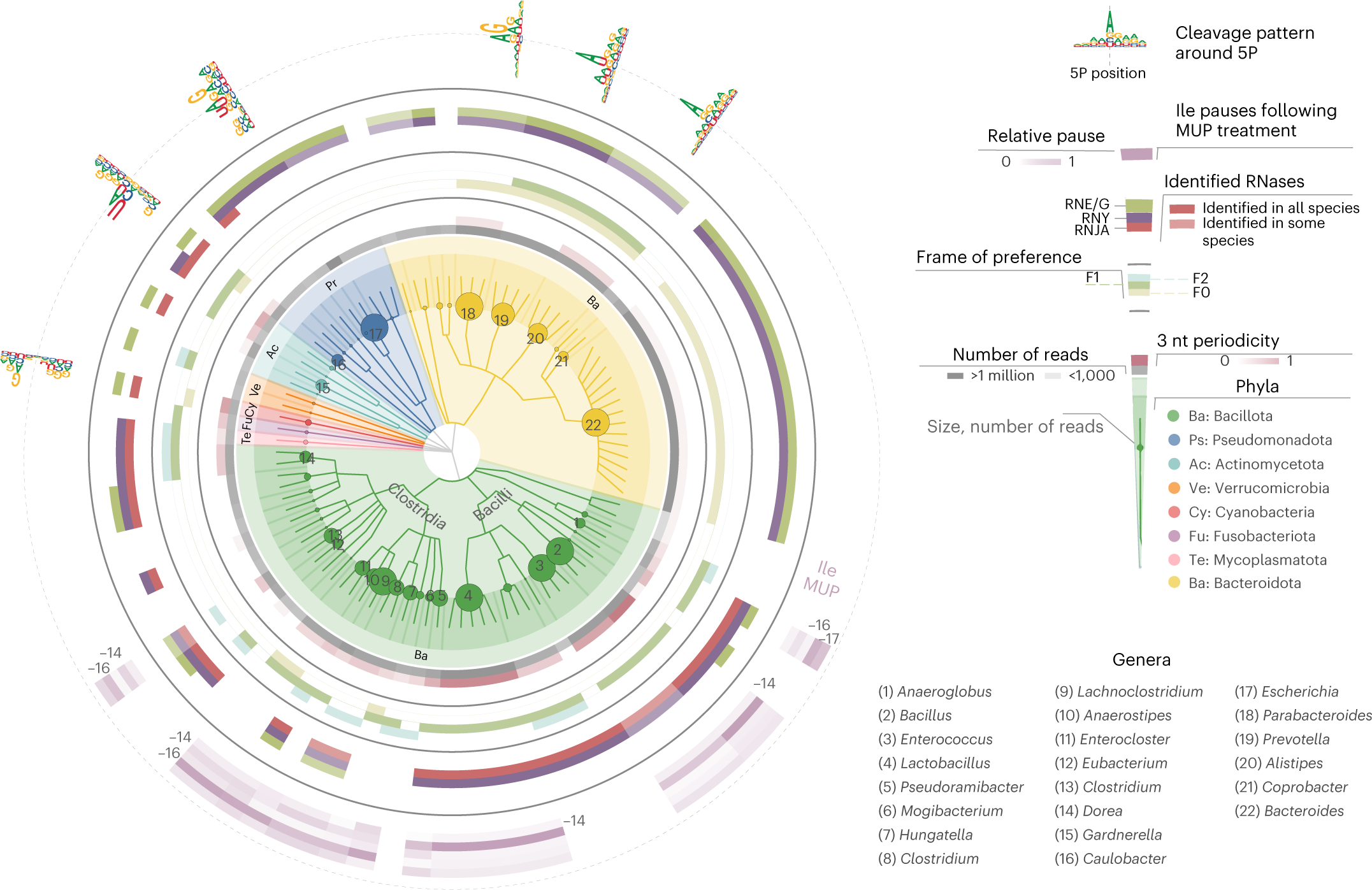
Co-translational mRNA decay is conserved across prokaryotes
Taxonomic tree of investigated species. Circles, from inside to outside, grey (number of assigned reads); red (strength of 3 nt periodicity), frame preference (F0, beige; F1, green; F2, light blue); identification of selected enzymes involved in RNA degradation at genus-level RNJA, RNY and RNE/G; ribonucleases from E/G protein family; accumulation of 5′P endpoints 13–17 nt before Ile codons after 30 min MUP treatment; cleavage motifs 4 nt around 5′P endpoints of selected species (skipping Bacillota with no cleavage preference). Overall, 58 genera were present in samples from cultured bacteria and complex environments, including vaginal swabs, faeces, faecal cultures and compost. In complex samples, species with at least 1,000 reads in the coding regions (300 in the case of compost) were considered, and respective genera analysed.
Examine how bacteria interact
Apart from measuring antibiotic resistance, the method can be used to help researchers understand how bacteria handle stress and interact with each other and with their hosts. The researchers will continue to study complex gut samples to examine how the bacterial communities interact in our gut and contribute to health and disease.
Source – Karolinska Institutet
Availability – an interactive browsable website for further exploration: http://metadegradome.pelechanolab.com
Huch S, Nersisyan L, Ropat M, Barrett D, Wu M, Wang J, Valeriano VD, Vardazaryan N, Huerta-Cepas J, Wei W, Du J, Steinmetz LM, Engstrand L, Pelechano V. (2023) Atlas of mRNA translation and decay for bacteria. Nat Microbiol [Epub ahead of print]. [article]