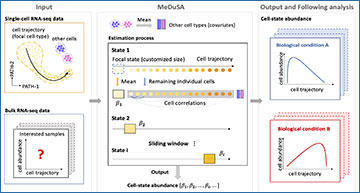
Deconvoluting cell-state abundances from bulk RNA-sequencing data can add considerable value to existing data, but achieving fine-resolution and high-accuracy deconvolution remains a challenge. Researchers at Zhejiang University have developed MeDuSA, a mixed model-based method that leverages single-cell RNA-sequencing data as a reference to estimate cell-state abundances along a one-dimensional trajectory in bulk RNA-sequencing data. The advantage of MeDuSA lies primarily in estimating cell abundance in each state while fitting the remaining cells of the same type individually as random effects. Extensive simulations and real-data benchmark analyses demonstrate that MeDuSA greatly improves the estimation accuracy over existing methods for one-dimensional trajectories. Applying MeDuSA to cohort-level RNA-sequencing datasets reveals associations of cell-state abundances with disease or treatment conditions and cell-state-dependent genetic control of transcription. This study provides a high-accuracy and fine-resolution method for cell-state deconvolution along a one-dimensional trajectory and demonstrates its utility in characterizing the dynamics of cell states in various biological processes.
MeDuSA is a fine-resolution cellular deconvolution method that leverages scRNA-seq data as a reference to estimate cell-state abundance along a one-dimensional trajectory in bulk RNA-seq data. MeDuSA features the use of a linear mixed model (LMM) to fit a cell state in question (either a single cell or the mean of multiple cells) as a fixed effect and the remaining cells of the same cell type individually as random effects accounting for correlations between cells. This model improves the deconvolution accuracy because the random-effect component allows each cell has a specific weight on bulk gene expression, resulting in a better capturing of variance in bulk gene expression. This model aslo ameliorates the collinearity problem between cells at the focal state (fitted as a fixed effect) and those at adjacent states (fitted as random effects) because of the shrinkage of random effects.
Availability – The source code of MeDuSA is available at https://github.com/LeonSong1995/MeDuSA
Song L, Sun X, Qi T, Yang J. (2023) Mixed model-based deconvolution of cell-state abundances (MeDuSA) along a one-dimensional trajectory. Nat Comput Sci [Epub ahead of print]. [article]