Detecting differentially expressed genes is important for characterizing subpopulations of cells. In scRNA-seq data, however, nuisance variation due to technical factors like sequencing depth and RNA capture efficiency obscures the underlying biological signal. Deep generative models have been extensively applied to scRNA-seq data, with a special focus on embedding cells into a low-dimensional latent space and correcting for batch effects. However, little attention has been paid to the problem of utilizing the uncertainty from the deep generative model for differential expression (DE). Furthermore, the existing approaches do not allow for controlling for effect size or the false discovery rate (FDR).
University of California, Berkeley researchers have developed lvm-DE, a generic Bayesian approach for performing DE predictions from a fitted deep generative model, while controlling the FDR. They apply the lvm-DE framework to scVI and scSphere, two deep generative models. The resulting approaches outperform state-of-the-art methods at estimating the log fold change in gene expression levels as well as detecting differentially expressed genes between subpopulations of cells.
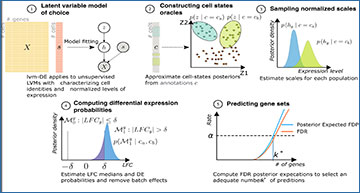
Differential expression model for deep generative models
(A) lvm-DE takes annotated data (from clustering, metadata, or transfer learning), a latent variable model, and a target FDR level as inputs and returns LFC estimates as well as calibrated DE predictions. (B) lvm-DE works as follows. 1) A preliminary step consists in fitting the latent variable model of choice of the data from the collection of available scRNA-seq data. 2) lvm-DE uses existing cell states annotations to approximate the distributions of c conditioned on the cell states. 3) These distributions help determine the normalized expression level distributions of the compared populations. 4) The associated LFC distribution helps to determine posterior DE probabilities that correspond to the model in which the LFC is higher than a given threshold. 5) To tag DE genes of interpretable interest, we estimate the maximum number of genes for which the posterior expected FDR is below the desired FDR level.
Availability – The implementation to reproduce the experiments of this paper and the reference implementation of scVI data have been deposited in Github (https://github.com/PierreBoyeau/lvm-DE-reproducibility and https://github.com/scverse/scvi-tools).
Boyeau P, Regier J, Gayoso A, Jordan MI, Lopez R, Yosef N. (2023) An empirical Bayes method for differential expression analysis of single cells with deep generative models. PNAS 120(21):e2209124120. [article]