Researchers from Osaka University use a novel combination of long- and short-read RNA sequencing to uncover the mechanism behind this biological mystery.
Daphnia are tiny crustaceans, or ‘water fleas’, that are extremely adaptable to their environment. This is owing to their remarkable phenotypic plasticity, i.e., their ability to change their form or behavior despite their genetic makeup remaining unchanged. Even male and female Daphnia are genetically identical!
Now, a group at Osaka University has shed some light on this phenomenon, by combining two different RNA sequencing techniques to reveal that certain genes produce different versions, or isoforms, of the proteins they encode in male and female Daphnia.
Phenotypic plasticity allows a single genome to produce different ‘phenotypes’, meaning Daphnia can physiologically adapt to variations in water salinity, temperature, the presence of predators, and more. A molecule called RNA is produced from a gene in a process called transcription, which provides instructions for the cellular machinery to then make the corresponding protein. Changes to the RNA after transcription can result in multiple isoforms, or slightly different RNA molecules, being produced from a single gene.
Such isoform variants are difficult to study because most RNA sequencing techniques read only very short stretches of RNA molecules, and then rely on computational techniques to piece the sequences back together. This means the fine details of different isoforms are lost.
However, in a novel approach, the team first used a newer, long-read RNA sequencing method called Iso-Seq to precisely identify all the different isoforms present in a species called Daphnia magna, before narrowing down the specific switching of isoforms occurring between males and females using highly accurate short-read RNA sequences.
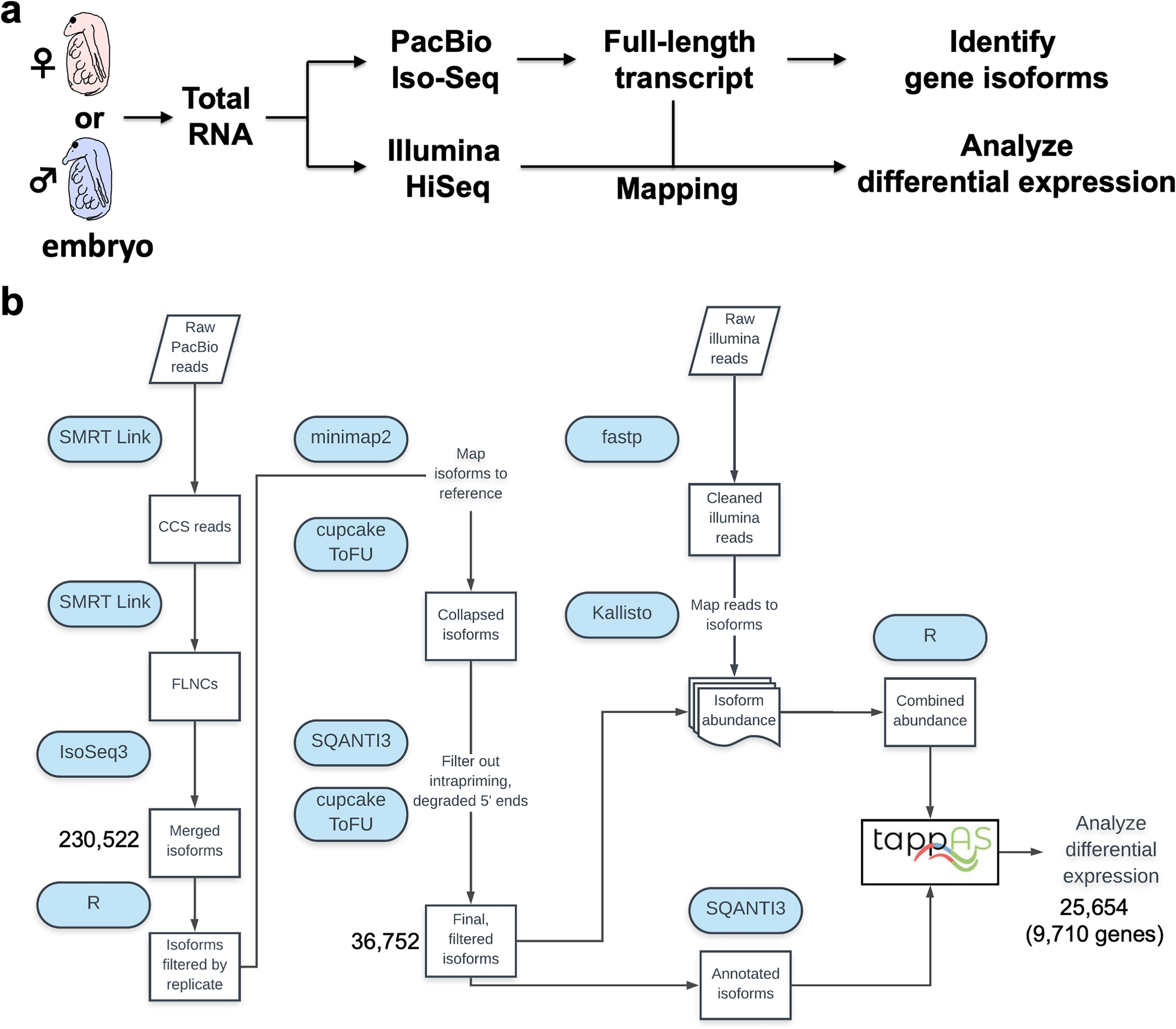
Daphnia embryo transcriptome workflow
(a) Overview of analysis in this study. Total RNAs were prepared from female or male embryos at 40 h after ovulation and divided into two samples each of which is subjected to PacBio Iso-Seq or Illumina HiSeq. Data of full-length transcripts from Iso-seq analysis not only led to the identification of gene isoforms but also was used for mapping the reads from Hiseq to analyze differential gene expression. (b) Workflow up to differential expression analysis in tappAS. CCS, circular consensus reads; FLNCs, full-length non-concatemers. The numbers of merged isoforms and final filtered isoforms were shown in addition to the numbers of isoforms and genes subjected to differential expression analysis.
“Short-read sequencing alone is unable to uncover this information,” explains first author Yasuhiko Kato. “This novel approach, combining long- and short-read sequencing, reveals previously unknown nuances of Daphnia transcription.”
“We identified genes that switch which specific isoforms are expressed in a sex-dependent manner,” adds senior author Hajime Watanabe. “This provides insight into the biological mechanisms that underlie the unique phenotypic plasticity of Daphnia.”
This research reveals the molecular basis for sexual dimorphism in Daphnia, an ecologically important species, and provides a tool for understanding how environmental conditions can determine sex in this species. These findings thus open the way for further advancements in crustacean aquaculture.
Source – Osaka University
Kato Y, Nitta JH, Perez CAG, Adhitama N, Religia P, Toyoda A, Iwasaki W, Watanabe H. (2024) Identification of gene isoforms and their switching events between male and female embryos of the parthenogenetic crustacean Daphnia magna. Sci Rep 14(1):9407. [article]