In experiments with significant perturbations to transcription, nascent RNA sequencing protocols are dependent on external spike-ins for reliable normalization. Unlike in RNA-seq, these spike-ins are not standardized and, in many cases, depend on a run-on reaction that is assumed to have constant efficiency across samples. To assess the validity of this assumption, University of Colorado researchers analyzed a large number of published nascent RNA spike-ins to quantify their variability across existing normalization methods. Furthermore, the researchers have developed a new biologically-informed Bayesian model to estimate the error in spike-in based normalization estimates, which they term Virtual Spike-In (VSI). They apply this method both to published external spike-ins as well as using reads at the 3′ end of long genes. The researchers find that spike-ins in existing nascent RNA experiments are typically under sequenced, with high variability between samples. Furthermore, they show that these high variability estimates can have significant downstream effects on analysis, complicating biological interpretations of results.
A Bayesian model describing normalization data for nascent RNA sequencing data
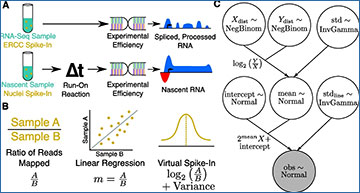
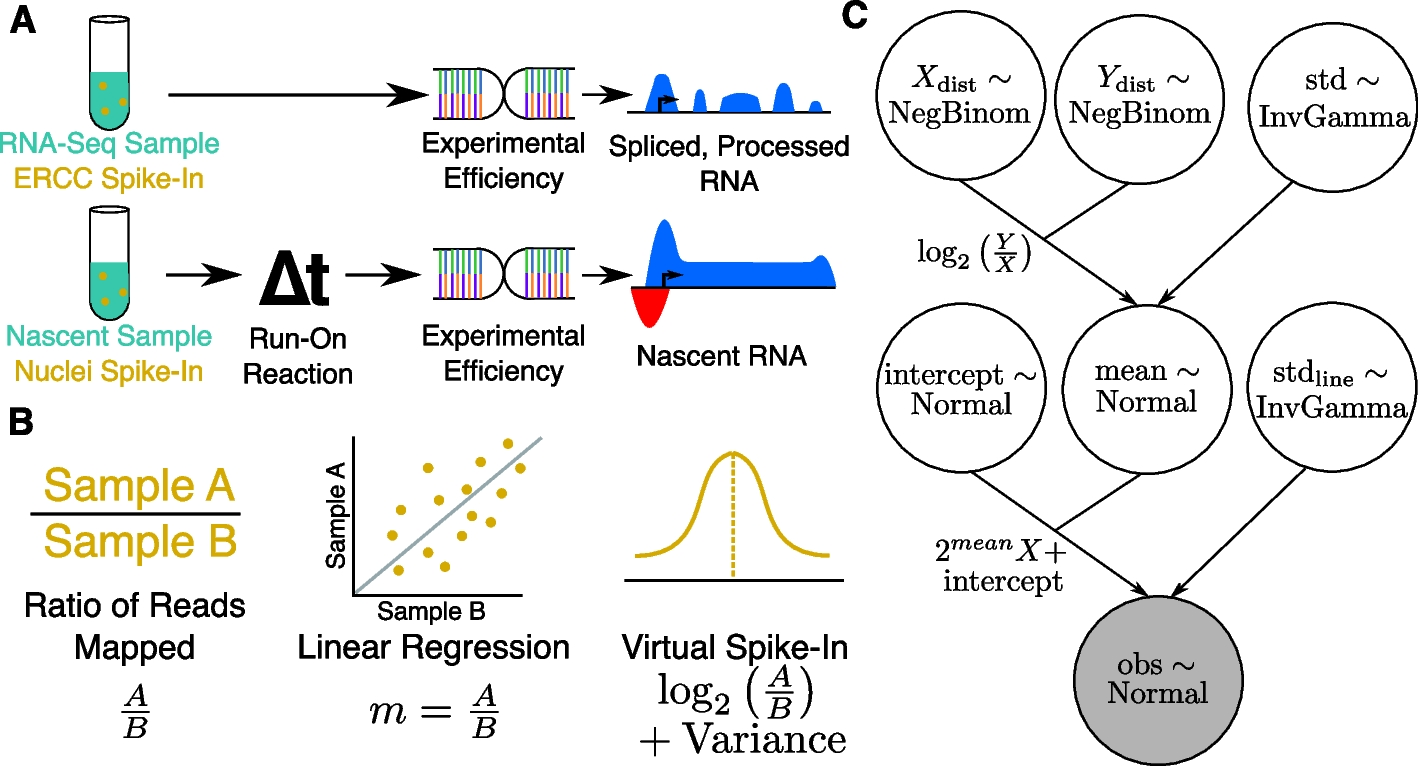
A Schematic showing typical external control, handling, and resulting data profile differences between RNA-seq (top) and run-on nascent RNA sequencing assays (bottom). Note that run-on efficiency is assumed to be equivalent between spike-in nuclei and experimental nuclei. B Quantifying a normalization factor is accomplished either by a naive ratio of total reads approach (left), linear regression (middle), or by the Bayesian model proposed here (right). Linear regression (middle) is more resistant to noise and outliers, but does not provide a reliable way to measure the variance of the normalization estimate. The Bayesian model (right) converts the slope to log space, converting the multiplicative nature of the normalization factor to a linear one, for which normalization factors can be readily inferred as a normal distribution with variance. C A plate diagram showing the VSI model as implemented in pymc3. Briefly, we estimate our count distributions X and Y (top row) with a negative binomial. The ratio of two negative binomial distributions is approximately log-normal, so we derive a normal distribution called mean (middle) as the log of the ratio of Y and X with some variance (top right), estimated as an inverse gamma distributed random variable. With the estimation of the mean established, we then add additional parameters to describe the intercept, and variance of the actual line of best fit. This is done so that the parameter mean is estimating an error in log-transformed space, as discussed in Panel (B)
Maas ZL, Dowell RD. (2024) Internal and external normalization of nascent RNA sequencing run-on experiments. BMC Bioinformatics 25(1):19. [article]