Expanding the clinical use of RNA sequencing improves diagnostic yield, reduces inconclusive rates, and improves health equity
Ambry Genetics, a prominent leader in clinical genomic testing and a subsidiary of REALM IDx, Inc., unveiled at the American Society of Human Genetics (ASHG) annual meeting results of a study published in JAMA Oncology that assessed the cumulative impact of paired DNA and RNA testing on detecting disease-causing germline genetic variants and resolution of variants of uncertain significance (VUS).
Key findings from this study indicate that RNA-dependent classifications were instrumental in enhancing the accuracy of genetic testing. Specifically, RNA sequencing played a pivotal role in:
- Improving Diagnostic Yield: Simultaneous RNA sequencing resulted in a significant increase in the diagnostic yield, providing more accurate and actionable information for individuals undergoing germline cancer predisposition testing.
- Reducing Inconclusive Rates: The number of variants of uncertain significance reduced considerably, providing clarity and enhancing the overall reliability of multi-gene panel testing.
- Improving Health Equity: Genetic cohorts are typically derived from European populations, resulting in evidence gaps in underrepresented populations. Generating novel functional evidence helps close those gaps via preferential improvement in VUS rates in non-white populations.
Furthermore, the study revealed that RNA sequencing had a significant impact on the classification of splicing variants. Specifically:
- Classification of 17.1% of Pathogenic Splicing Variants Dependent on RNA evidence: 17.1% of splicing variants classified as pathogenic would have been classified as variants of uncertain significance without RNA evidence and would therefore not have been clinically actionable.
- 71.1% of Splicing Variants of Uncertain Significance (VUS) Resolved as Benign: RNA sequencing resolved 71.1% of splicing variants initially categorized as VUS, reducing ambiguity and enhancing the accuracy of hereditary cancer testing.
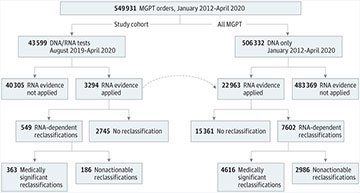
Sankey Diagrams Depicting Change in Variant Classifications Based on RNA Evidence
Comparisons of variant classifications at the onset of the study before RNA evidence was available (left side of diagrams) with variant classifications after RNA evidence was applied (right side of diagrams). A, Change in variant classifications based on RNA evidence in all genes together. B, Change in variant classifications based on RNA evidence in BRCA1 and BRCA2. C, Change in variant classifications based on RNA evidence in other breast/ovarian genes: BRIP1, CDH1, CHEK2, NF1, PTEN, PALB2, RAD51C, RAD51D, and TP53. D, Change in variant classifications based on RNA evidence in mismatch repair and polyposis genes: APC, MLH1, MSH2, MSH6, MUTYH, and PMS2.
These findings signify a shift in the field of cancer predisposition, particularly towards the utilization of multiomics genetic testing, providing individuals with more reliable and actionable information, without sacrificing turnaround time. This technology enables healthcare providers to support their patients in making personalized, timely, evidenced-based medical management decisions. As the field of genetic testing continues to evolve, RNA sequencing promises to play a pivotal role in advancing precision medicine.
“The integration of DNA with RNA sequencing into germline cancer predisposition evaluation is the new gold standard for genetic testing,” states Rachid Karam, M.D., Ph.D., VP of Research & Development at Ambry Genetics and lead investigator of this study. “We’ve witnessed a remarkable improvement in diagnostic accuracy and a substantial reduction in inconclusive results. Patients benefit from having access to the most comprehensive genetic testing possible, as demonstrated by the thousands of individuals impacted by RNA testing in our laboratory cohort alone.”
Source – Ambry Genetics
Horton C, Hoang L, Zimmermann H, Young C, Grzybowski J, Durda K, Vuong H, Burks D, Cass A, LaDuca H, Richardson ME, Harrison S, Chao EC, Karam R. (2023) Diagnostic Outcomes of Concurrent DNA and RNA Sequencing in Individuals Undergoing Hereditary Cancer Testing. JAMA Oncol [Epub ahead of print]. [article]