Rare cell types can have an undue influence on human health. Previous research has suggested that a subset of astrocytes—star-shaped cells in the brain and spinal cord—may be responsible for multiple sclerosis (MS), a disease in which the immune system attacks the covering that protects nerves. But finding these rare cells is no easy task—to pinpoint them, investigators need to identify unique surface markers that can distinguish these culprit cells from others.
Single-cell RNA sequencing can help find them, even in the absence of distinguishing surface marker, but this technique can become extremely expensive. To address this problem, a team led by investigators from Brigham and Women’s Hospital, a founding member of the Mass General Brigham healthcare system, developed FIND-seq, which combines nucleic acid cytometry, microfluidics, and droplet sorting to isolate and analyze rare cells of interest based on the expression of mRNA biomarkers detected by digital droplet PCR. Using this method, the team analyzed in great detail a population of astrocytes that drives central nervous system inflammation and neurodegeneration. When used in combination with other tools, FIND-seq identified signaling pathways controlled by the mineralocorticoid receptor NR3C2 and the nuclear receptor corepressor 2 that play important roles in the development of pathogenic astrocytes in mice and humans. In another study, researchers used FIND-seq to identify mechanisms used by HIV to “hide” in immune cells in patients treated with anti-retroviral therapies.
Whole-transcriptomic analysis of HIV-DNA+ cells using FIND-seq
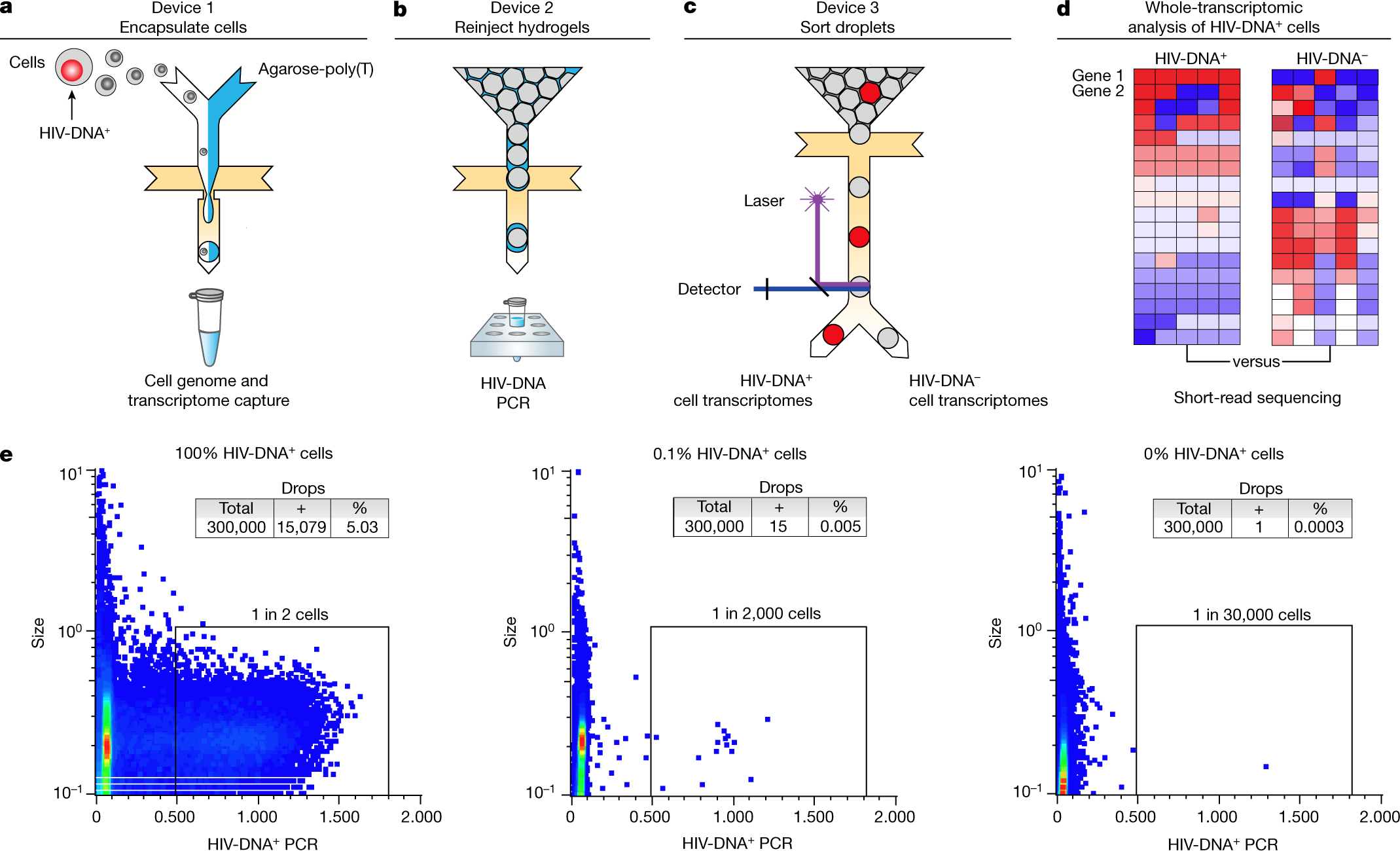
a–d, Overview of the workflow, including three sequential microfluidic devices separated by handling steps. a, On the first device, single cells are encapsulated at a limiting dilution in a water-in-oil emulsion with lysis buffer and molten agarose-poly(T). The agarose is then cooled to form a hydrogel bead that retains genomic DNA and polyadenylated RNA. After oil removal, whole-transcriptome cDNA is covalently linked to the hydrogel by reverse transcription for subsequent whole-transcriptome amplification (WTA) using PCR. b,c, Hydrogel beads re-encapsulated on the second device are analysed using droplet PCR for HIV gag (b) and then sorted on the third device (c). d, The processing steps performed after droplet sorting include WTA, library preparation and sequencing, and bioinformatic comparison of HIV-DNA+ cells and HIV-DNA− cells. e, Droplet cytometry plots demonstrating the analysis of pure HIV-DNA+ J-Lat T cells (left), pure HIV-DNA− Jurkat T cells (right), and a mixture of 0.1% J-Lat and 99.9% Jurkat cells (middle). Cells were encapsulated at 1 cell per 10 droplets.
“These findings identify novel targets for therapeutic intervention in neurologic diseases such as MS,” said corresponding author Francisco Quintana, PhD, of the BWH Department of Neurology. The team is working to develop novel small molecules which could be used to target this pathway therapeutically.
Source – Brigham and Women’s Hospital
Clark IC, Wheeler MA, Lee HG, Li Z, Sanmarco LM, Thaploo S, Polonio CM, Shin SW, Scalisi G, Henry AR, Rone JM, Giovannoni F, Charabati M, Akl CF, Aleman DM, Zandee SEJ, Prat A, Douek DC, Boritz EA, Quintana FJ, Abate AR. (2023) Identification of astrocyte regulators by nucleic acid cytometry. Nature [Epub ahead of print]. [abstract]
Clark IC, Mudvari P, Thaploo S, Smith S, Abu-Laban M, Hamouda M, Theberge M, Shah S, Ko SH, Pérez L, Bunis DG, Lee JS, Kilam D, Zakaria S, Choi S, Darko S, Henry AR, Wheeler MA, Hoh R, Butrus S, Deeks SG, Quintana FJ, Douek DC, Abate AR, Boritz EA. (2023) HIV silencing and cell survival signatures in infected T cell reservoirs. Nature [Epub ahead of print]. [article]