Spatially resolved transcriptomic technologies show promise in revealing complex pathophysiological processes, but developing sensitive, high-resolution, and cost-effective methodology is challenging. Researchers at the Shanghai Jiao Tong University School of Medicine have developed a dendrimeric DNA coordinate barcoding design for spatial RNA sequencing (Decoder-seq). This technology combined dendrimeric nano-substrates with microfluidic coordinate barcoding to generate high-density spatial DNA arrays with deterministically combinatorial barcodes in a resolution-flexible and cost-effective manner (~$0.5/mm2). Decoder-seq achieved high RNA capture efficiency, ~68.9% that of in situ sequencing, and enhanced the detection of lowly expressed genes by ~five-fold compared to 10× Visium. Decoder-seq visualized a spatial single-cell atlas of mouse hippocampus at near-cellular resolution (15 μm) and revealed dendrite-enriched mRNAs. Application to renal cancers dissected the heterogeneous tumor microenvironment of two subtypes, and identified spatial gradient expressed genes with the potential in predicting tumor prognosis and progression. Decoder-seq is compatible with sensitivity, resolution, and cost, making spatial transcriptomic analysis accessible to wider biomedical applications and researchers.
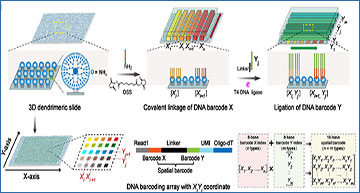
(a) Generation of 3D dendrimeric DNA coordinate barcoding array. 3D dendrimeric nano-substrates offer multiple active primary amino groups to allow high-density DNA modification. A pair of microchannel chips consecutively introduce two sets of DNA barcodes to dendrimeric nano-substrates, allowing covalent linkage of DNA barcode X and ligation of DNA barcode Y to form XiYj coordinates at the intersections. The linker consists of linker 1 and linker 2. Linker 1 from 3’ DNA barcode X while linker 2 from 5’ DNA barcode Y. Thus, a DNA barcoding array with n × m unique XiYj coordinates was formed with only n + m unique DNA barcodes. (b) Spatial transcriptome indexing. In situ capture and RT of mRNAs from tissue sections were conducted on DNA coordinate barcoding arrays, resulting in cDNAs for library preparation and sequencing. (c) Spatially resolved transcriptomics analysis. A spatially-resolved transcriptomics map was reconstructed by integrating gene expression data with the spatial barcodes via bioinformatic analysis.
Cao J, Zheng Z, Sun D, Chen X, Chen R, Lv T, An Y, Zheng J, Song J, Wu L, Yang C. (2023) Dendrimeric DNA Coordinate Barcoding Design for Spatial RNA Sequencing. bioRXiv [online preprint]. [abstract]