Element Biosciences researchers present avidity sequencing, a sequencing chemistry that separately optimizes the processes of stepping along a DNA template and that of identifying each nucleotide within the template. Nucleotide identification uses multivalent nucleotide ligands on dye-labeled cores to form polymerase-polymer-nucleotide complexes bound to clonal copies of DNA targets. These polymer-nucleotide substrates, termed avidites, decrease the required concentration of reporting nucleotides from micromolar to nanomolar and yield negligible dissociation rates. Avidity sequencing chemistry enables a diversity of applications that include single-cell RNA sequencing (RNA-seq) and whole-human-genome sequencing. Avidity sequencing achieves high accuracy, with 96.2% and 85.4% of base calls having an average of one error per 1,000 and 10,000 base pairs, respectively. The researchers show that the average error rate of avidity sequencing remained stable following a long homopolymer.
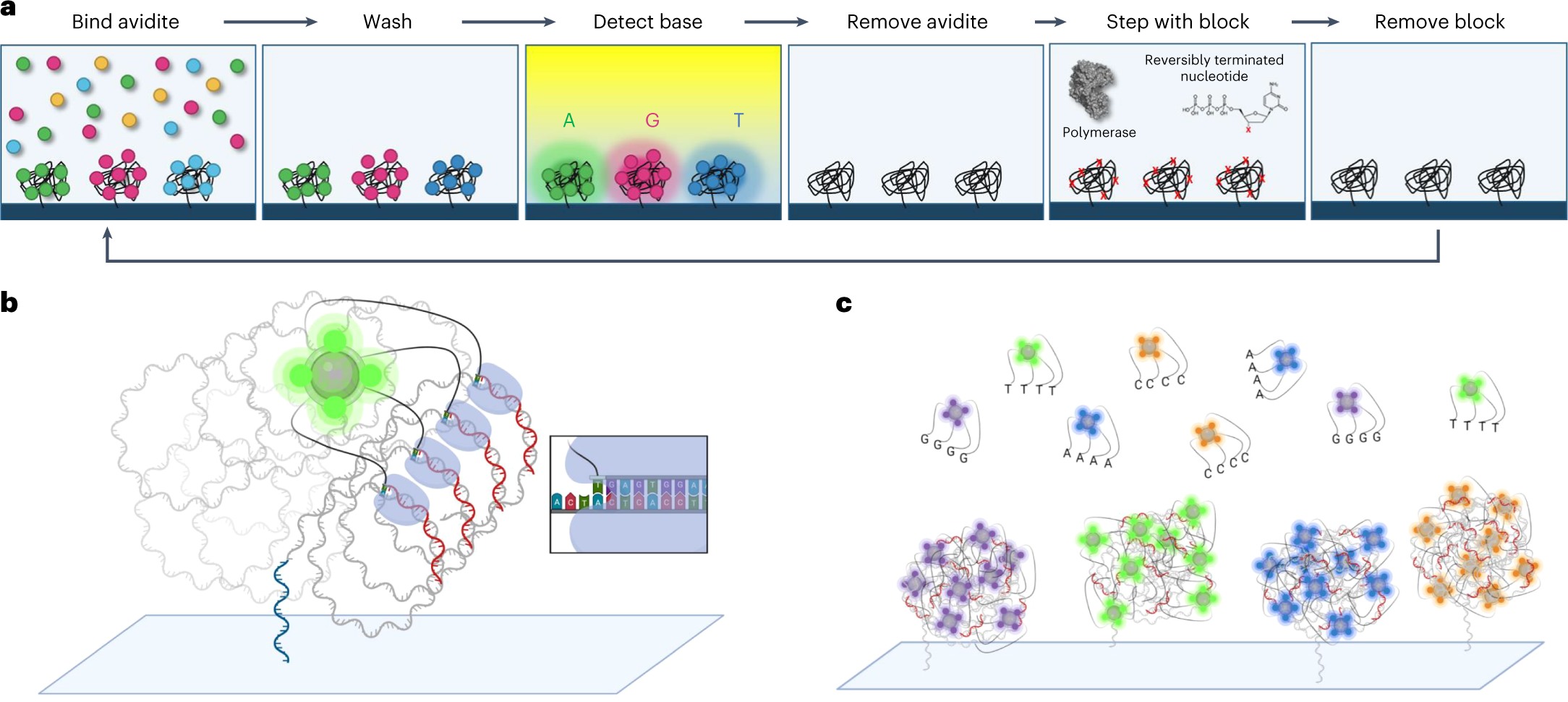
Avidity sequencing workflow and scheme
a, Sequencing by avidity. A reagent containing multivalent avidite substrates and an engineered polymerase are combined with DNA polonies inside a flowcell. The engineered polymerase binds to the free 3′ ends of the primer-template of a polony and selects the correct cognate avidite via base-pairing discrimination. The multivalent avidite interacts with multiple polymerases on one polony to create avidity binding that reduces the effective Kd of the avidite substrates 100-fold compared with a monovalent dye-labeled nucleotide, allowing productive binding of nanomolar concentrations. Multiple polymerase-mediated binding events per avidite ensure a long signal persistence time. Imaging of fluorescent, bound avidites enables base classification. Following detection, avidites are removed from the polonies. Extension by one base using an engineered polymerase incorporates an unlabeled, blocked nucleotide. A terminal 3′ hydroxyl is regenerated on the DNA strand, allowing repetition of the cycle. b, Rendering of a single avidite bound to a DNA polony via polymerase-mediated selection. The initial surface primer used for library hybridization and extension during polony formation is shown in blue. Sequencing primers (red) are shown annealed to the single-strand DNA polony (gray). Each arm of the avidite (black) connects the avidite core containing multiple fluorophores (green) to a nucleotide substrate. The polymerase bound to the sequencing primer selects the correct nucleotide to base pair with the templating base (inset). The result is multiple base-mediated anchor points noncovalently attaching the avidite to the DNA polony. c, Rendering of multiple DNA polonies with template-specific avidites bound during the binding step of the cycle (polymerase not shown for simplicity). Many avidites bind to each DNA polony generating a fluorescent signal during detection. Multiple long, flexible polymer linkers connect the core to the nucleotide substrates.
Arslan S, Garcia FJ, Guo M, Kellinger MW, Kruglyak S, LeVieux JA, Mah AH, Wang H, Zhao J, Zhou C, Altomare A, Bailey J, Byrne MB, Chang C, Chen SX, Cho B, Dennler CN, Dien VT, Fuller D, Kelley R, Khandan O, Klein MG, Kim M, Lajoie BR, Lin B, Liu Y, Lopez T, Mains PT, Price AD, Robertson SR, Taylor-Weiner H, Tippana R, Tomaney AB, Zhang S, Abtahi M, Ambroso MR, Bajari R, Bellizzi AM, Benitez CB, Berard DR, Berti L, Blease KN, Blum AP, Boddicker AM, Bondar L, Brown C, Bui CA, Calleja-Aguirre J, Cappa K, Chan J, Chang VW, Charov K, Chen X, Constandse RM, Damron W, Dawood M, DeBuono N, Dimalanta JD, Edoli L, Elango K, Faustino N, Feng C, Ferrari M, Frankie K, Fries A, Galloway A, Gavrila V, Gemmen GJ, Ghadiali J, Ghorbani A, Goddard LA, Guetter AR, Hendricks GL, Hentschel J, Honigfort DJ, Hsieh YT, Hwang Fu YH, Im SK, Jin C, Kabu S, Kincade DE, Levy S, Li Y, Liang VK, Light WH, Lipsher JB, Liu TL, Long G, Ma R, Mailloux JM, Mandla KA, Martinez AR, Mass M, McKean DT, Meron M, Miller EA, Moh CS, Moore RK, Moreno J, Neysmith JM, Niman CS, Nunez JM, Ojeda MT, Ortiz SE, Owens J, Piland G, Proctor DJ, Purba JB, Ray M, Rong D, Saade VM, Saha S, Tomas GS, Scheidler N, Sirajudeen LH, Snow S, Stengel G, Stinson R, Stone MJ, Sundseth KJ, Thai E, Thompson CJ, Tjioe M, Trejo CL, Trieger G, Truong DN, Tse B, Voiles B, Vuong H, Wong JC, Wu CT, Yu H, Yu Y, Yu M, Zhang X, Zhao D, Zheng G, He M, Previte M. (2023) Sequencing by avidity enables high accuracy with low reagent consumption. Nat Biotechnol [Epub ahead of print]. [article]