The application of single-cell RNA sequencing (scRNA-seq) in biomedical research has advanced our understanding of the pathogenesis of disease and provided valuable insights into new diagnostic and therapeutic strategies. With the expansion of capacity for high-throughput scRNA-seq, including clinical samples, the analysis of these huge volumes of data has become a daunting prospect for researchers entering this field.
Researchers at Nanjing Medical University review the workflow for typical scRNA-seq data analysis, covering raw data processing and quality control, basic data analysis applicable for almost all scRNA-seq data sets, and advanced data analysis that should be tailored to specific scientific questions. While summarizing the current methods for each analysis step, the researchers also provide an online repository of software and wrapped-up scripts to support the implementation. Recommendations and caveats are pointed out for some specific analysis tasks and approaches. The researchers hope this resource will be helpful to researchers engaging with scRNA-seq, in particular for emerging clinical applications.
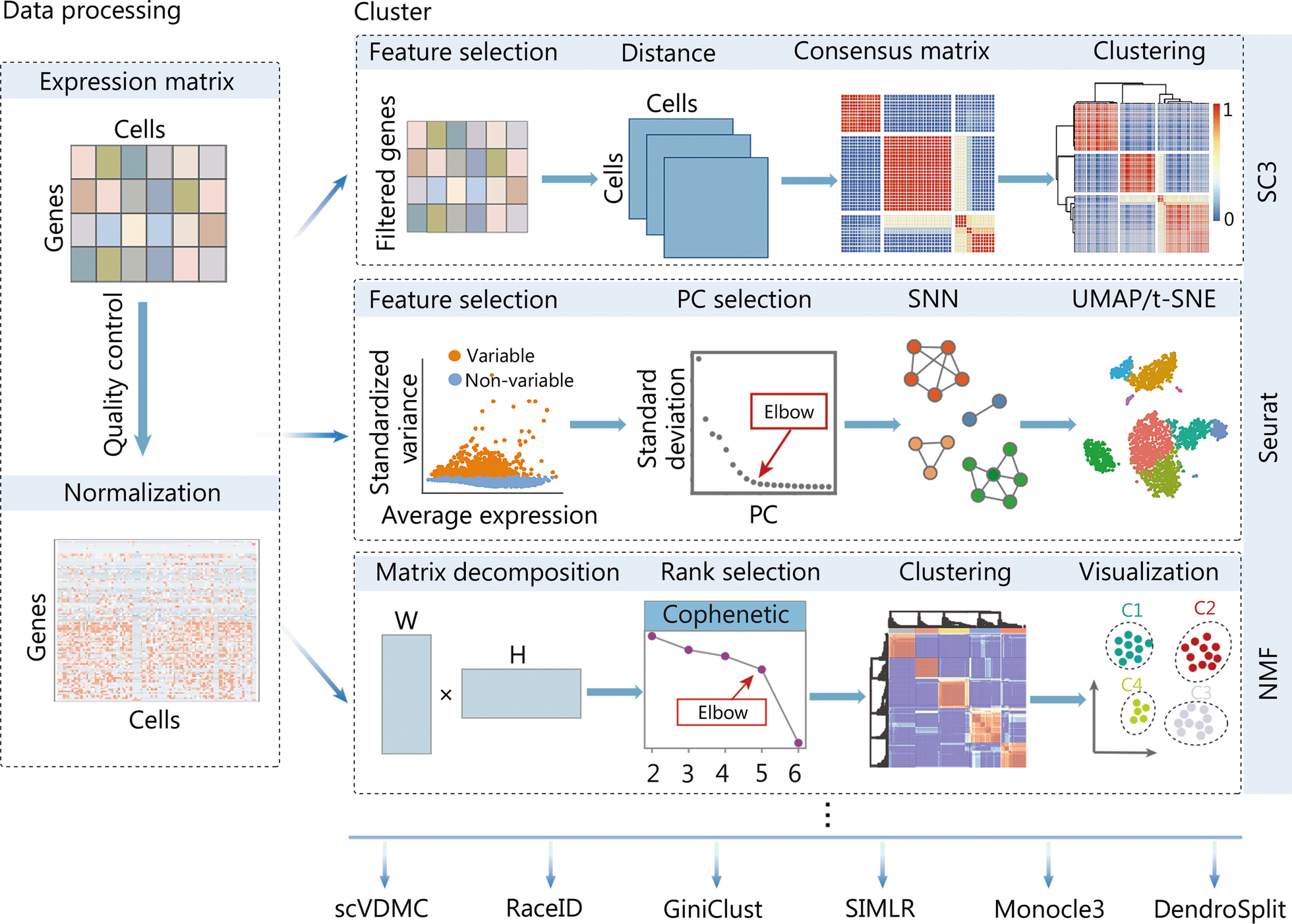
Typical computational strategies and methods for clustering cells using scRNA-seq data
With the processed scRNA-seq data, the SC3 approach, the Seurat clustering implementation based on the community detection method, and the NMF method are popular choices. scRNA-seq single-cell RNA sequencing, SC3 single-cell consensus clustering, NMF non-negative matrix factorization, PC principal component, SNN shared nearest neighbor, scVDMC variance-driven multitask clustering of scRNA-seq data, SIMLR single-cell interpretation via multikernel learning, UMAP uniform manifold approximation and projection, t-SNE t-distributed stochastic neighbor embedding
Su M, Pan T, Chen QZ, Zhou WW, Gong Y, Xu G, Yan HY, Li S, Shi QZ, Zhang Y, He X, Jiang CJ, Fan SC, Li X, Cairns MJ, Wang X, Li YS. (2022) Data analysis guidelines for single-cell RNA-seq in biomedical studies and clinical applications. Mil Med Res 9(1):68. [article]