The human genome has just over 20,000 genes coding for proteins. Yet, it produces at least ten times that many different non-coding RNA molecules, which can often take on more than one shape. At least some of this RNA structurome is functional in physiology or pathophysiology. In an invited review for Nature Reviews Genetics, Danny Incarnato, a molecular geneticist from the University of Groningen (The Netherlands), and his colleague Robert C. Spitale from the University of California, Irvine in California (USA) describe ways to develop the, as yet, largely untapped potential of RNA structures.
RNA is perhaps best known as the intermediate between genome and protein synthesis: messenger RNA molecules copy the genetic code of a gene in the cell’s nucleus and transport it to the cytoplasm, where ribosomes translate the code into a protein. However, RNA is also a key regulator of almost every cellular process and the structures that are adopted by RNA molecules are thought to often be key to their functions.
Function
Danny Incarnato, Assistant Professor of Molecular Genetics, has long been interested in the role of RNA structures in the cell and works on methods to elucidate the different structures in living cells. So, when he was invited to write a review on RNA structures, he accepted without hesitation.
‘And I was happy to invite my friend and colleague Robert Spitale, one of the pioneers of the ‘RNA revolution’, to join me.’
In recent years, the knowledge of RNA molecules in the cell has increased dramatically. The ENCODE project revealed the huge number of non-coding RNAs in cells; in human cells, over ten times higher than the number of coding genes.
‘Not all of them have a function,’ Incarnato stresses. ‘But many do, and with regard to their variety, we have barely scratched the surface.’
Switches
Different types of non-coding RNAs have been known for a long time and it was also clear that their structure could play an important role. An example are riboswitches: RNAs that can respond to changes in the external environment by changing shape, which in turn can affect specific gene activity.
‘We also knew that RNA molecules can act as enzymes,’ says Incarnato. ‘And, of course, ribosomes are RNA structures.’
Thus, RNA molecules can act as sensors, catalysts, switches, or scaffolds and affect RNA translation, but can also affect RNA degradation and alternative splicing.
Drug research
It is, therefore, no surprise that RNAs have rapidly gained momentum in drug research. Yet, our knowledge of the ‘structurome’ is still very limited.
‘So far, we have almost exclusively looked at single structures. But RNA molecules are very dynamic and molecules with the same sequence can take on different shapes,’ explains Incarnato. ‘Due to the way these structures were determined, they are often averages of all the possible conformations of a single molecule.’
Read out of high-throughput sequencing (HTS)-based RNA structure mapping experiments
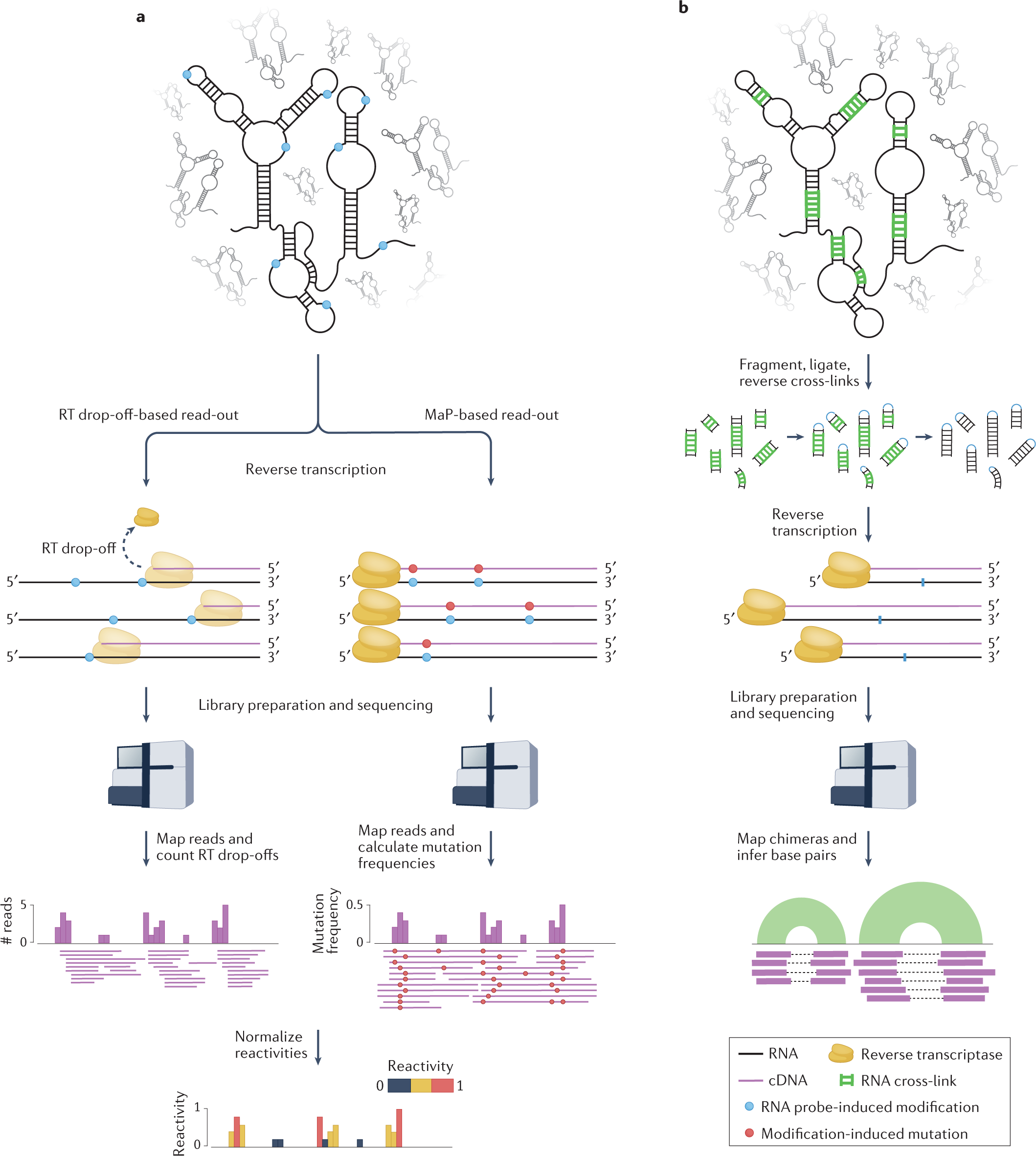
a, In chemical probing experiments, RNA undergoes reverse transcription following treatment with the chemical probe. When drop-off-based read-outs are used, the reverse transcriptase (RT) drops off the template at sites that have reacted with the probe, resulting in a pool of truncated cDNA molecules that terminate at the nucleotide prior to the modified site. Alternatively, in mutational profiling (MaP) experiments, reverse transcription conditions are adjusted so that the RT reads through the chemically modified sites but incorporates incorrect bases, resulting in (possibly full-length) cDNAs containing mutations at modification sites. In both cases, cDNA fragments are ligated to adapters, converted to double-stranded DNA libraries and sequenced. Sequencing reads (corresponding to cDNA fragments) are mapped back to the reference transcriptome. For RT drop-off-based experiments, each position i along the RNA is assigned a count corresponding to the number of reads whose 5′ ends mapped one nucleotide downstream (i + 1). For MaP-based experiments, the mutation frequency at each position of the RNA is calculated as the ratio between the number of mutated reads and the total number of reads covering that position. These raw reactivity profiles are then normalized to yield reactivities ranging between 0 (unreactive) and, depending on the normalization method, ≥1 (highly reactive). b, In direct RNA–RNA interaction capture experiments, RNA duplexes are cross-linked (for example by psoralen), RNA is fragmented and the two strands of the cross-linked duplexes are intramolecularly ligated, after which cross-linking is reversed. These chimeric RNA fragments are then reverse-transcribed and the resulting cDNA fragments are ligated to adapters, converted to double-stranded DNA libraries and sequenced. Sequencing reads are then mapped back to the reference transcriptome. As these reads are derived from RNA chimeras, the two halves of these reads will map to distinct locations of the same transcript in the case of intramolecular duplexes, or distinct transcripts in the case of intermolecular duplexes.
RNA viruses
Incarnato has pioneered methods to uncover the structural heterogeneity of RNA molecules.
‘We can combine this with high throughput RNA-sequencing to probe the structural heterogeneity.’
In some cases, different structures are just an ’evolutionary by-product’, while in other cases they are functional.
Incarnato: ‘In this way, RNA molecules can regulate practically anything inside a cell and, therefore, play a role in both physiology and pathophysiology.’
Although the developments in this field are rapid, they don’t proceed in a very orderly manner.
Incarnato: ‘There is applied pharmaceutical research going on alongside lots of fundamental research.’ Interfering with RNAs could be an important way to fight diseases, including those caused by RNA viruses such as SARS-CoV2. ‘Yet, we don’t have a clue about off-target effects. For small molecules that interfere with specific enzymes, such as kinases, profiling panels are available to evaluate off-target profiles. However, we don’t know how many RNAs have similar shapes. We really need a clear map of the RNA structurome.’
SARS-CoV-2 genome: A shapeshifting killer. The RNA genome of the SARS-CoV-2 virus can adopt multiple ‘shapes’ (structural conformations) in the context of the infected host cell. Although the function of the different conformations is still largely unknown, some of them have been shown to be crucial during the virus replicative cycle, for example to regulate genome cyclization. | Illustraion Danny Incarnato, University of Groningen
Software
Another problem is that in many cases, it is impossible to know which of the different structural versions of a given RNA molecule is responsible for its function or dysfunction.
’And on top of that, RNAs can interact and create complicated regulatory networks. So, we also need a deeper understanding of how this works in cells.’
There is a lot of work still to do. In addition, software is important; computer programs are needed to translate biochemical analyses of RNAs into their different structures.
‘In our field, you need to know as much about coding as about high throughput sequencing. All of us are at home both in wet labs and in bioinformatics.’
Source – University of Groningen
Spitale RC, Incarnato D. (2022) Probing the dynamic RNA structurome and its functions. Nat Rev Genet [Epub ahead of print]. [article]