Microbial communities are massively resident in the human body, yet dysbiosis has been reported to correlate with many diseases, including various cancers. Most studies focus on the gut microbiome, while the bacteria that participate in tumor microenvironments on site remain unclear. Previous studies have acquired the bacteria expression profiles from RNA-seq, whole genome sequencing, and whole exon sequencing in The Cancer Genome Atlas (TCGA). However, small-RNA sequencing data were rarely used. Using TCGA miRNA sequencing data, researchers at the National Taiwan University evaluated bacterial abundance in 32 types of cancer. To uncover the bacteria involved in cancer, the researchers applied an analytical process to align unmapped human reads to bacterial references and developed the BIC database for the transcriptional landscape of bacteria in cancer. BIC provides cancer-associated bacterial information, including the relative abundance of bacteria, bacterial diversity, associations with clinical relevance, the co-expression network of bacteria and human genes, and their associated biological functions. These results can complement previously published databases. Users can easily download the result plots and tables, or download the bacterial abundance matrix for further analyses. In summary, BIC can provide information on cancer microenvironments related to microbial communities.
BIC user interface and analysis modules
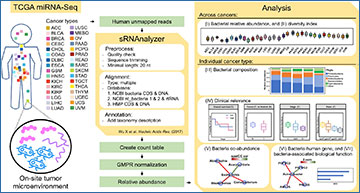
(A) The user interface and analysis modules. (B, C) Modules I and II show the relative abundance and diversity of bacteria across cancers. (D) Module III shows the bacteria composition. (E–H) Module IV shows the clinical relevance, such as overall survival, and relative abundance compared in different groups (tumor versus adjacency normal; tumor stages; races). (I) Module V shows the bacteria co-abundance network. (J) Module VI shows the bacteria-correlated human gene expression network. (K) Module VII shows the bacteria-associated biological functions.
Availability – BIC is available at: http://bic.jhlab.tw/.
Chen KP, Hsu CL, Oyang YJ, Huang HC, Juan HF. (2022) BIC: a database for the transcriptional landscape of bacteria in cancer. Nucleic Acids Res [Epub ahead of print]. [article]