The rapid development of single cell transcriptomics has revolutionized the study of complex tissues. Single cell RNA-sequencing (scRNA-seq) can profile tens-of-thousands of dissociated cells from a single tissue sample, enabling researchers to identify the cell types, phenotypes and interactions that control tissue structure and function. A key requirement for these applications is the accurate estimation of cell surface protein abundance. Although technologies to directly quantify surface proteins are available, this data is uncommon and limited to proteins with available antibodies. While supervised methods that are trained on both transcriptomics and proteomics data (e.g., Cellular Indexing of Transcriptomes and Epitopes by Sequencing or CITE-seq) can provide the best performance, this training data is also limited by available antibodies and may not exist for the tissue under investigation. In the absence of protein measurements, researchers must estimate receptor abundance from scRNA-seq data.
Researchers at Dartmouth College have developed a new unsupervised method for receptor abundance estimation using scRNA-seq data, Surface Protein abundance Estimation using CKmeans-based clustered thresholding (SPECK), and evaluated its performance against other unsupervised approaches on up to 215 human receptors and multiple tissue types. This analysis reveals that techniques based on a thresholded reduced rank reconstructed (RRR) output of scRNA-seq data are effective for receptor abundance estimation with the SPECK method providing the best overall performance.
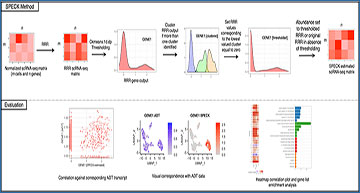
SPECK Method
SPECK performs normalization, rank selection, reduced rank reconstruction and thresholding on a m × n scRNA-seq count matrix with m cells and n genes. The reconstructed and thresholded matrix is of size m × n. To evaluate SPECK, receptor abundance estimates were visually assessed using feature plots and heat maps and correspondence to CITE-seq data was quantified using the Spearman rank correlation.
Availability – Current R package implementation for SPECK is available on the Comprehensive R Archive Network (CRAN): https://cran.r-project.org/web/packages/SPECK/index.html
Javaid A, Frost HR. (2022) SPECK: An Unsupervised Learning Approach for Cell Surface Receptor Abundance Estimation for Single Cell RNA-Sequencing Data. bioRXiv [online preprint]. [abstract]