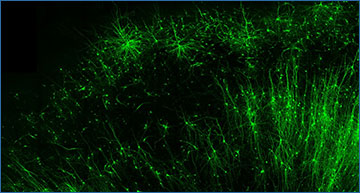
Tagging and illuminating only the inhibitory “brake” cells (green) in human brain tissue is just one of many things the new tool, CellREADR, can do. Credit – Derek Southwell, Duke University
Duke University researchers have developed an RNA-based editing tool that targets individual cells, rather than genes. It is capable of precisely targeting any type of cell and selectively adding any protein of interest.
Researchers said the tool could enable modifying very specific cells and cell functions to manage disease.
Using an RNA-based probe, a team led by neurobiologist Z. Josh Huang, Ph.D. and postdoctoral researcher Yongjun Qian, Ph.D. demonstrated they can introduce into cells fluorescent tags to label specific types of brain tissue; a light-sensitive on/off switch to silence or activate neurons of their choosing; and even a self-destruct enzyme to precisely expunge some cells but not others. The work appears Oct. 5 in Nature.
Their selective cell monitoring and control system relies on the ADAR enzyme, which is found in every animal’s cells. While these are early days for CellREADR (Cell access through RNA sensing by Endogenous ADAR), the possible applications appear to be endless, Huang said, as is its potential to work across the animal kingdom.
“We’re excited because this provides a simplified, scalable and generalizable technology to monitor and manipulate all cell types in any animal,” Huang said. “We could actually modify specific types of cell function to manage diseases, regardless of their initial genetic predisposition,” he said. “That’s not possible with current therapies or medicine.”
CellREADR is a customizable string of RNA made up of three main sections: a sensor, a stop sign, and a set of blueprints.
First, the research team decides what specific cell type they want to investigate, and identifies a target RNA that is uniquely produced by that cell type. The tool’s remarkable tissue specificity relies on the fact that each cell type manufactures signature RNA not seen in other cell types.
A sensor sequence is then designed as the target RNA’s complementary strand. Just as the rungs on DNA are made-up of complementary molecules that are inherently attracted to each other, RNA has the same magnetic potential to link with another piece of RNA if it has matching molecules.
After a sensor makes its way into a cell and finds its target RNA sequence, both pieces glom together to create a piece of double-stranded RNA. This new RNA mashup triggers the enzyme ADAR to inspect the new creation and then change a single nucleotide of its code.
The ADAR enzyme is a cell-defense mechanism designed to edit double-strand RNA when it occurs, and is believed to be found in all animal cells.
Knowing this, Qian designed CellREADR’s stop sign using the same specific nucleotide ADAR edits in double-stranded RNA. The stop sign, which prevents the protein blueprints from being built, is only removed once CellREADR’s sensor docks to its target RNA sequence, making it highly specific for a given cell type.
Once ADAR removes the stop sign, the blueprints can be read by cellular machinery that builds the new protein inside the target cell.
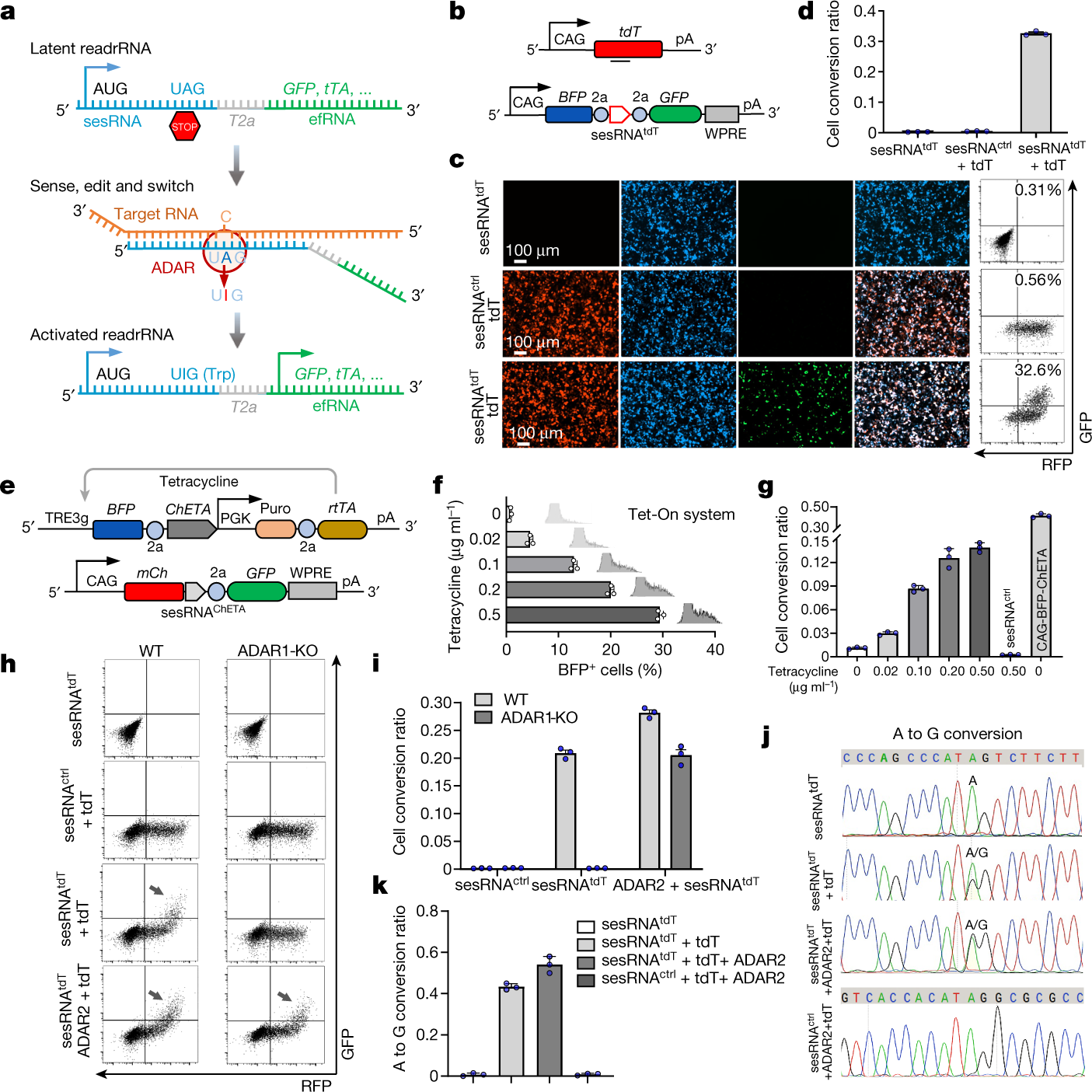
CellREADR design and implementation in mammalian cells
a, Schematic showing the design of CellREADR. CellREADR is a modular readrRNA molecule, consisting of a 5′ sensor domain (sesRNA) and 3′ effector domain (efRNA), separated by a T2A coding region. sesRNA is complementary to a cellular RNA and contains a stop codon that prevents efRNA translation. Base-pairing between sesRNA and target RNA recruits ADARs, which mediate A-to-I editing and convert the UAG stop to a UGG Trp codon, switching on translation of effector protein. b, CAG-tdT encodes tdTomato target RNA and READRtdT-GFP encodes a readrRNA consisting of a BFP sequence followed by sesRNAtdT and efRNAGFP. WPRE and 2a are sequences for a virus post-transcriptional regulatory element and a self-cleaving peptide coding sequence, respectively. pA, polyadenylated tail. c, Cells transfected with both CAG-tdT (tdT) and READRtdT-GFP exhibited robust GFP expression that co-localized with BFP and RFP (bottom). Cells transduced with READRtdT-GFP only (top) or CAG-tdT and READRctrl encoding sesRNActrl (middle) exhibited very low GFP expression. Right, FACS analysis of GFP and RFP expression. The percentage of GFP+ cells is indicated. d, Quantification of CellREADR efficiency by FACS analysis. e–g, The effect of target RNA expression levels on CellREADR. e, rtTA-TRE3g-ChETA (top) is designed so that BFP-ChETA target RNA is transcribed from TRE3g, driven by constitutively expressed rtTA in a tetracycline-concentration-dependent manner. READRChETA-GFP (bottom) is designed to express readrRNA comprising mCherry (mCh) followed by sesRNAChETA and efRNAGFP. f, Increasing tetracycline concentrations resulted in increasing numbers of BFP+ cells, expressed here as a percentage of RFP+ cells (conversion ratio). g, The conversion ratio increased with increasing tetracycline concentration. CAG-ChETA, which results in constitutive expression of BFP-ChETA, served as positive control. READRctrl served as negative control. h,i, ADAR1 is required for CellREADR function. FACS analysis (h) and quantification (i) of wild-type (WT) or ADAR1-KO cells with sesRNAtdT only, sesRNActrl and CAG-tdT, sesRNAtdT and CAG-tdT, or sesRNAtdT and CAG-tdT with overexpression of ADAR2. Arrows indicate GFP+RFP+ populations. j,k, Electropherograms of Sanger sequencing (j) and quantification (k), showing A to G conversion at the intended editing site in different samples. Data in d,f,g,i,k are mean ± s.e.m.; n = 3 independent experiments.
In their paper, Huang and his team put CellREADR through its paces.
“I remember two years ago when Yongjun built the first iteration of CellREADR and tested it in a mouse brain,” Huang said. “To my amazement, it worked spectacularly on his first try.”
The team’s careful planning and design paid off as they were then able to demonstrate CellREADR accurately labelled specific brain cell populations in living mice, as well as effectively added activity monitors and control switches where directed. It also worked well in rats, and in human brain tissue collected from epilepsy surgeries.
“With CellREADR, we can pick and choose populations to study and really begin to investigate the full range of cell types present in the human brain,” said co-author Derek Southwell, M.D., Ph.D., a neurosurgeon and assistant professor in the department of neurosurgery at Duke.
Southwell hopes CellREADR will improve his and others’ understanding of the wiring diagram for human brain circuits and the cells within them, and in doing so, help advance new therapies for neurological disorders, such as a promising new method to treat drug-resistant epilepsy he is piloting.
Huang and Qian are especially hopeful about CellREADR’s potential as a “programmable RNA medicine” to possibly cure diseases — since that’s what drew them both to science in the first place. They have applied for a patent on the technology.
“When I majored in pharmacology as an undergraduate, I was very naïve,” Qian said. “I thought you could do a lot of things, like cure cancer, but actually it’s very difficult. However, now I think, yes maybe we can do it.”
Source – Duke University
Qian Y, Li J, Zhao S, Matthews EA, Adoff M, Zhong W, An X, Yeo M, Park C, Yang X, Wang BS, Southwell DG, Huang ZJ. (2022) Programmable RNA sensing for cell monitoring and manipulation. Nature [Epub ahead of print]. [abstract]