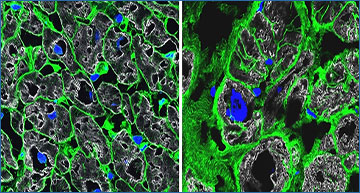
Microscope images of healthy heart tissue (left) and diseased heart tissue (right). Cell boundaries are stained in green, nuclei in blue, and heart muscle in gray. Images: Anissa Viveiros and Gavin Oudit/University of Alberta
Heart failure is a common and devastating disorder for which there is no cure. Many conditions that make it difficult for the heart to pump blood—such as dilated cardiomyopathy and arrhythmogenic cardiomyopathy—can lead to heart failure, but treatments for patients with heart failure do not take these distinct conditions into account.
Investigators from Harvard Medical School and Brigham and Women’s Hospital set out to identify molecules and pathways that may contribute to heart failure, with the aim of informing more effective and personalized treatments.
Using single nucleus RNA sequencing, or snRNAseq, to gain insight into the specific changes that occur in different cell types and cell states, the team made several surprising discoveries.
They found that while there are some shared genetic signatures, others are distinct, providing new candidate targets for therapy and predicting that personalized treatment could improve patient care. Results were published online August 4 in Science.
Genotype-stratified analyses of heart failure at the single-nuclei level

The transcriptomes of 881,081 nuclei from 61 heart failure patients were profiled and compared with the transcriptional signatures of 18 healthy controls. Genotype-stratified analyses of cell types and cell state compositions, differential gene expression, cell-cell interactions, and machine learning illuminated the shared and distinct transcriptional signatures resulting from pathogenic variants in DCM and ACM.
“Our findings hold enormous potential for rethinking how we treat heart failure and point to the importance of understanding its root causes and the mutations that lead to changes that may alter how the heart functions,” said co-senior author Christine Seidman, the Thomas W. Smith Professor of Medicine and professor of genetics in the Blavatnik Institute at HMS and director of the Cardiovascular Genetics Center at Brigham and Women’s.
“This is fundamental research, but it identifies targets that can be experimentally pursued to propel future therapeutics,” she said. “Our findings also point to the importance of genotyping. Not only does genotyping empower research but it can also lead to better, personalized treatment for patients.”
Team effort

An echocardiogram shows abnormal heart structures and function—notably enlarged left atria and ventricle and reduced contraction in the ventricle—in a patient with dilated cardiomyopathy. Video: Brigham and Women‘s
Seidman and Jonathan Seidman, the Henrietta B. and Frederick H. Bugher Foundation Professor of Genetics at HMS, collaborated with an international team.
To conduct their study, the Seidmans and colleagues analyzed samples from 18 control and 61 failing human hearts from patients with dilated cardiomyopathy, arrhythmogenic cardiomyopathy, or an unknown cardiomyopathy disease.
The human heart is composed of many different cell types, including cardiomyocytes (beating heart cells), fibroblasts (which help form connective tissue and contribute to scarring), and smooth muscle cells. The team used single nucleus RNA sequencing to look at the genetic readouts from individual cells and determine cellular and molecular changes in each distinct cell type.
from STAT News
Looking at RNA molecules gives us a snapshot of how much a gene is active or inactive at a particular time point. Until recently, we couldn’t do that in the heart because the approach had been to take heart tissue, grind it all up, and look at the RNAs that are up or down. But that gives you what we call a smoothie: It’s all the different component cells — those strawberries, blueberries, bananas — mixed together.
But there’s a technology now called single-cell RNA sequencing. And that says, what are the RNAs that are up or down in the cardiomyocytes as compared to the smooth muscle cells, as compared to the fibroblasts, all of which are in the cells? You get a much more precise look at what’s changing in a different cell type. And that’s the approach that we use, because cardiomyocytes [the cells in the heart that make it contract]are very large. They’re at least three times bigger than other cells. We can’t capture the single cell — it literally does not fit through the microfluidic device. And so we sequenced the nuclei, which is where the RNA emanates from.
From these data, the team identified 10 major cell types and 71 distinct transcriptional states.
They found that in the tissue from patients with dilated or arrhythmogenic cardiomyopathy, cardiomyocytes were depleted while endothelial and immune cells were increased. Overall, fibroblasts did not increase but showed altered activity.
Analyses of multiple hearts with mutations in certain disease genes—including TTN, PKP2, and LMNA—uncovered molecular and cellular differences as well as some shared responses.
The team also leveraged machine learning approaches to identify cell and genotype patterns in the data. This approach further confirmed that while some disease pathways converged, differences in genotype promoted distinct signals, even in advanced disease.
The authors note that future studies are needed to further define the molecular underpinnings of cardiomyopathies and heart failure across sex, age, and other demographics as well as across different areas of the heart. The team has made its datasets and platform freely available.
“We could not have done this work without sample donations from patients,” said Christine Seidman. “Our goal is to honor their contributions by accelerating research and making our work available so that others can continue to advance what we understand about disease, improve treatment, and work on strategies to prevent heart failure.”
Source – Brigham and Women’s Hospital
Reichart D, Lindberg EL, Maatz H, Miranda AMA, Viveiros A, Shvetsov N, Gärtner A, Nadelmann ER, Lee M, Kanemaru K, Ruiz-Orera J, Strohmenger V, DeLaughter DM, Patone G, Zhang H, Woehler A, Lippert C, Kim Y, Adami E, Gorham JM, Barnett SN, Brown K, Buchan RJ, Chowdhury RA, Constantinou C, Cranley J, Felkin LE, Fox H, Ghauri A, Gummert J, Kanda M, Li R, Mach L, McDonough B, Samari S, Shahriaran F, Yapp C, Stanasiuk C, Theotokis PI, Theis FJ, van den Bogaerdt A, Wakimoto H, Ware JS, Worth CL, Barton PJR, Lee YA, Teichmann SA, Milting H, Noseda M, Oudit GY, Heinig M, Seidman JG, Hubner N, Seidman CE. (2022) Pathogenic variants damage cell composition and single cell transcription in cardiomyopathies. Science 377(6606):eabo1984. [article]