From Intel by Sanchit Misra
There are many kinds of single-cell analyses studying various aspects of cell-differentiation. Single-cell RNA-seq (scRNA-seq) analysis studies the differences in gene expression profiles across cells. It relies on single-cell RNA sequencing, which is an advanced technique that enables measurement of the gene expression of individual cells.
A typical workflow to do scRNA-seq analysis begins with a matrix that consists of the expression levels of the genes in each cell. In the data preprocessing steps, noise is filtered out and the data is normalized to obtain the activity of every human gene in each individual cell of the dataset. During this step, machine learning is often utilized to correct artifacts from data collection. Subsequently, dimensionality reduction is performed followed by clustering to group cells with similar genetic activity and visualization of the clusters. With over 800,000 downloads, Scanpy is one of the most widely used toolkits for this analysis.
Pipeline showing the steps in analysis of single-cell RNA sequencing data starting from gene activity matrix to visualization of different cell clusters.
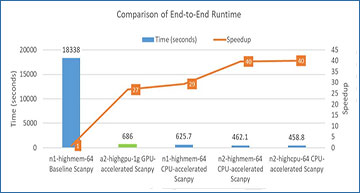
For a dataset consisting of 1.3 million mouse brain cells, the pipeline depicted above in Figure 1 would normally take nearly 5 hours on a single CPU instance (n1-highmem-64) on GCP using off-the-shelf (baseline) Scanpy implementation. For the same pipeline, Nvidia has reported an end-to-end runtime of 686 seconds on a single A100 GPU using Nvidia RAPIDS.