Researchers from Monash University have demonstrated how genetic variation influences the expression of genes that encode proteins involved in critical immune regulatory and signalling pathways in a cell type-specific manner.
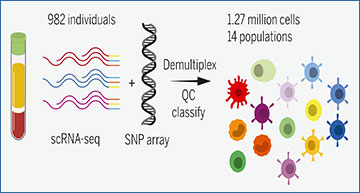
The researchers present single-cell RNA sequencing (scRNA-seq) data from 1.27 million peripheral blood mononuclear cells collected from 982 donors. They developed a framework for the classification of individual cells, and by combining the single-cell RNA sequencing data with genotype data, mapped the genetic effects on gene expression in each of 14 immune cell types.
If the genome is an architect’s blueprint, RNA sequencing of the transcriptome is the completed housing development. Single-cell RNA sequencing then enables us to look into individual houses on that lot.
“The study brings together population genetics and single-cell RNA sequencing data to uncover drivers of inter-individual variation in the immune system and the cell type-specific outcomes of gene expression. Understanding the genetic underpinnings of immune system regulation will eventually have broad implications in the treatment of autoimmune disease, infections, transplantation, and cancer,” Dr Rowson said.
Single-cell eQTL mapping and colocalization with autoimmune disease risk loci
scRNA-seq data from 1.27 million PBMCs were used to identify 26,597 cis-eQTLs (gray box). Dynamic eQTLs were uncovered as cells move from a naïve to a memory state (top right). Genetic variation between individuals influences immune regulation in a cell type–specific manner (middle right). In this study, 990 trans-eQTL effects (bottom right) and the causal effects for 305 autoimmune disease loci were identified (bottom left). Browsable results are available at www.onek1k.org. CD4NC, CD4 naïve and central memory T cells; CD8NC, CD8 naïve and central memory T cells; QC, quality control.
The study is the first large-scale cohort to undergo single-cell rather than bulk RNA sequencing, which provides a more nuanced indication of the specific cell types involved in gene expression, rather than assessing whole-of-immune system outcomes. It will enable the development of more targeted therapeutics for a host of diseases in future.
Source – Monash University
Yazar S, Alquicira-Hernandez J, Wing K, Senabouth A, Gordon MG, Andersen S, Lu Q, Rowson A, Taylor TRP, Clarke L, Maccora K, Chen C, Cook AL, Ye CJ, Fairfax KA, Hewitt AW, Powell JE. (2022) Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease. Science 376(6589):eabf3041. [abstract]