In the quest to understand gene regulation and cellular functions across different species, scientists have increasingly turned to a powerful technique known as single-cell RNA sequencing (scRNA-seq). This method allows researchers to examine the gene expression profiles of individual cells, providing a detailed view of cellular functions and interactions. However, analyzing and comparing scRNA-seq data across species to identify conserved genetic and cellular mechanisms has been a significant challenge—until now.
Introducing CACIMAR
Researchers at the Guangzhou Institutes of Biomedicine and Health have developed CACIMAR, a groundbreaking tool designed to perform cross-species analysis of cell identities, markers, regulations, and interactions using scRNA-seq data. CACIMAR stands for Cross-species Analysis of Cell Identities, Markers, and Regulations. This innovative tool enables researchers to uncover both molecular and cellular conservations, which are crucial for understanding evolutionary biology.
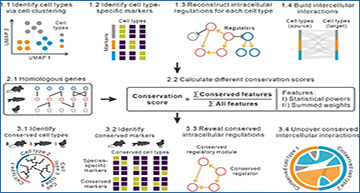
Schematics of CACIMAR
The scRNA-seq data from multiple species are used as the inputs of CACIMAR. First, CACIMAR identifies cell types, cell type-specific markers, intracellular regulations, and intercellular interactions using scRNA-seq data from each species. Second, CACIMAR calculates conservation scores based on homolog information of features. Features include cell types, markers, intracellular regulations, and intercellular interactions, some of which are represented by statistical powers or summed weights. Lastly, based on conservation score, CACIMAR allows to identify evolutionarily conserved cell types, markers, intracellular regulations, and intercellular interactions
Key Features of CACIMAR
CACIMAR leverages weighted sum models of conserved features to create different conservation scores. These scores measure the conservation of cell types, regulatory networks, and intercellular interactions. Here’s how CACIMAR can revolutionize our understanding of evolution:
- Identifying Conserved Cell Types: CACIMAR can identify cell types that are conserved across even evolutionarily distant species. This means that researchers can determine which cell types have remained relatively unchanged throughout evolution, providing insights into their fundamental roles.
- Marker Gene Identification: The tool helps identify genes that serve as markers for specific cell types, distinguishing between those that are conserved across species and those that are unique to a particular species.
- Intracellular Regulations: CACIMAR enables the identification of conserved regulatory networks within cells. This includes pinpointing specific regulatory subnetworks and key regulators that control gene expression in a cell type-specific manner.
- Intercellular Interactions: A unique feature of CACIMAR is its ability to identify conserved interactions between different cell types. Understanding these interactions is crucial for comprehending how cells communicate and coordinate their functions in different organisms.
Real-World Application: Retinal Regeneration
To showcase its capabilities, CACIMAR was tested using publicly available scRNA-seq data on retinal regeneration in mice, zebrafish, and chicks. Here’s what the tool achieved:
- Conserved Cell Types: Identified cell types involved in retinal regeneration that are conserved across these species.
- Marker Genes: Distinguished marker genes that are either conserved or specific to each species.
- Regulatory Networks: Revealed conserved regulatory mechanisms that control gene expression during retinal regeneration.
- Cell Interactions: Uncovered conserved interactions between different cell types involved in the regeneration process.
Why This Matters
Understanding which cellular and molecular mechanisms are conserved across species can shed light on fundamental biological processes and how they have evolved. This knowledge can lead to the discovery of crucial regulators and potential therapeutic targets for various diseases. For example, insights gained from studying retinal regeneration in animals could inform new treatments for eye injuries and diseases in humans.
Conclusion
CACIMAR represents a significant advancement in the field of evolutionary biology and transcriptomics. By facilitating the cross-species analysis of scRNA-seq data, it provides researchers with a powerful tool to uncover conserved cellular and molecular mechanisms. This, in turn, enhances our understanding of the evolutionary history of species and the fundamental processes that drive life.
Jiang J, Li J, Huang S, Jiang F, Liang Y, Xu X, Wang J. (2024) CACIMAR: cross-species analysis of cell identities, markers, regulations, and interactions using single-cell RNA sequencing data. Brief Bioinform 25(4):bbae283. [article]