In the world of biology, understanding how genes are regulated and how they interact within cells is crucial. One way scientists study these interactions is through single-cell RNA sequencing (scRNA-seq), which provides detailed information about gene expression in individual cells. However, studying these processes directly in living cells can be challenging and expensive. This is where computational tools like GRouNdGAN come into play.
Researchers at McGill University have developed GRouNdGAN, a sophisticated computational model designed to simulate single-cell RNA-seq data. But it does more than just create data; it also helps researchers understand how genes are regulated within cells. The “GRN” in GRouNdGAN stands for “Gene Regulatory Network,” which is a map showing how different genes control each other’s expression through various regulators, especially transcription factors (TFs).
GRouNdGAN generates data that mimic the real expression levels of genes in cells. It captures both steady-state (constant conditions) and transient-state (changing conditions) expressions. By incorporating user-defined gene regulatory networks, researchers can specify which genes regulate which others. This allows the model to simulate how changes in one gene’s activity can affect others. Moreover, the model preserves crucial features of real biological data, such as gene identities (which genes are present), cell trajectories (how cells progress over time), pseudo-time ordering (a way to arrange cells in a sequence based on their gene expression), and the inherent noise found in biological experiments.
Architecture and training procedure of GRouNdGAN
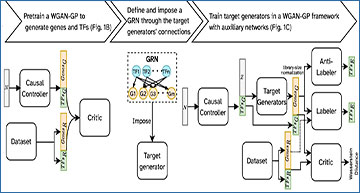
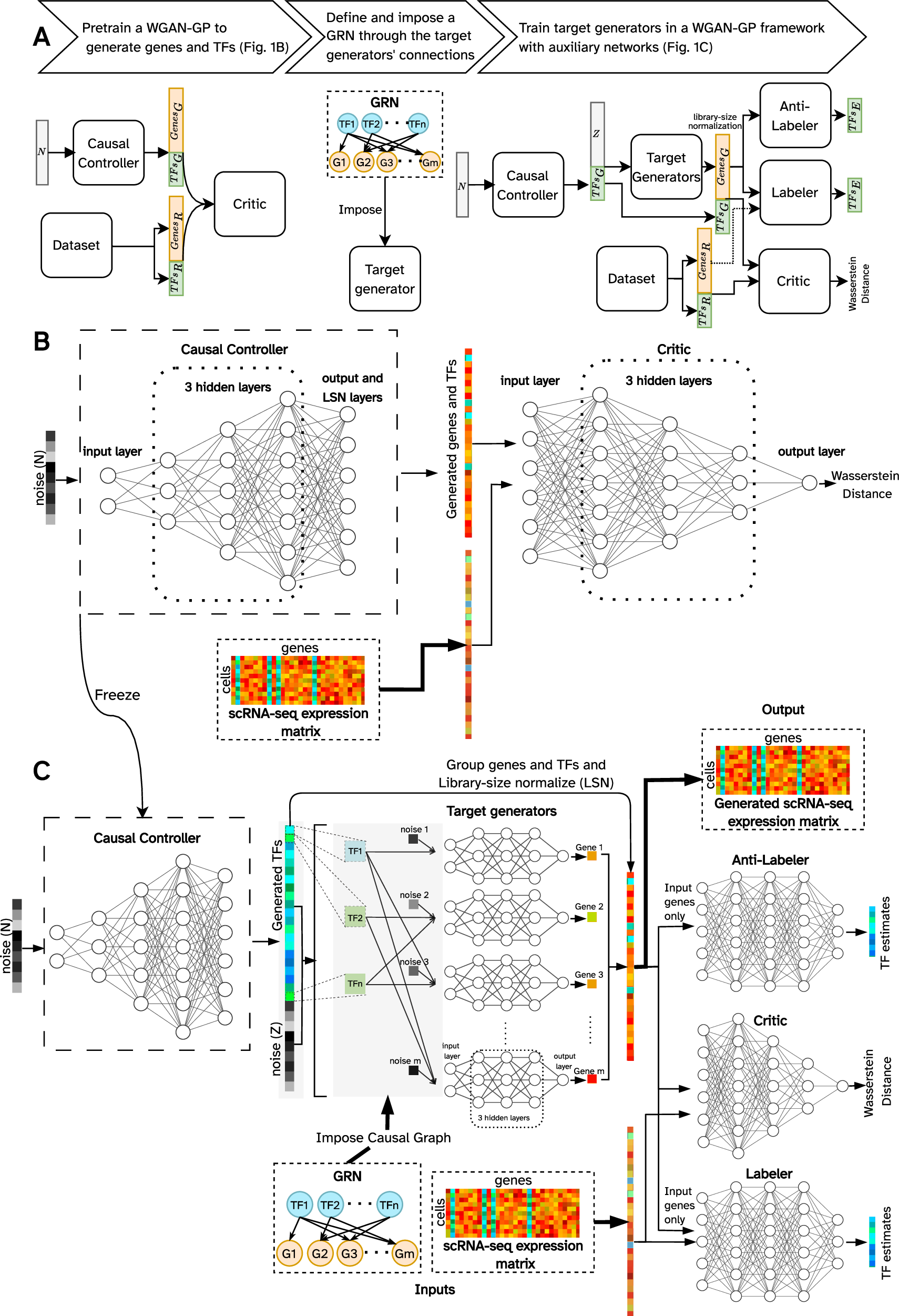
A The flowchart representing the steps of the training procedure and the overall architecture of the model in each step. Subscripts G, R, and E represent generated, real, and estimated, respectively. B A WGAN-GP is pre-trained to generate realistic simulated cells using the reference (real) training set. C The LSN layer of the generator of the trained WGAN-GP (panel B) is removed, its weights are frozen, and is used as the causal controller to generate unnormalized TF expression values (expression of target genes generated by the causal controller are discarded). These TF expression values along with a noise vector are provided as input to the target generators, following the provided causal GRN. The generated gene and TF expression values are reorganized and passed through the LSN layer. The normalized simulated expression vectors and experimental reference data (the same training set as B are then passed to the critic to estimate Wasserstein distance between the reference and the generated data distributions. The anti-labeler estimates TF values based on generated target gene expressions. The labeler performs a similar task, but in addition to receiving generated values, it also utilizes target gene expression values from the reference data. Labeler and anti-labeler ensure that the causal GRN is incorporated by the target generators. Details of the model are provided in Methods.
Researchers can use GRouNdGAN to perform virtual experiments, such as “knocking out” (disabling) specific transcription factors to see how other genes react. This is much like conducting lab experiments but in a computer. One major challenge in gene regulation research is the difference between simulated data and real biological data. GRouNdGAN helps bridge this gap by providing highly realistic simulations based on real experimental datasets. This makes it easier to test and validate gene regulatory network inference methods (ways to deduce the regulatory relationships between genes).
GRouNdGAN serves as a benchmark for testing other computational methods. By providing a known ground truth (the actual regulatory relationships it was programmed with), researchers can see how well their methods for inferring gene regulatory networks perform. Simulating experiments with GRouNdGAN can save considerable time and resources compared to conducting all experiments in a lab. This allows for rapid hypothesis testing and method development.
The applications of GRouNdGAN in real-world research are vast. In disease research, understanding gene regulatory networks is vital for studying diseases where gene expression is disrupted, such as cancer. GRouNdGAN can help simulate these conditions and test potential interventions. In drug development, by predicting how cells respond to different genetic modifications, GRouNdGAN can aid in identifying new drug targets and testing drug effects before moving to costly lab experiments. Basic biological research also benefits, as researchers studying fundamental questions about cell development, differentiation, and function can use GRouNdGAN to explore these processes in depth.
In summary, GRouNdGAN represents a powerful tool for the biological research community, allowing for detailed and realistic simulations of gene expression and regulation. By providing a bridge between computational and experimental data, it holds great promise for advancing our understanding of gene regulatory networks and their roles in health and disease.
Availability – GRouNdGAN along with a tutorial is freely available under the GNU Affero General Public License v3.0 on GitHub (https://github.com/Emad-COMBINE-lab/GRouNdGAN)
Zinati Y, Takiddeen A, Emad A. (2024) GRouNdGAN: GRN-guided simulation of single-cell RNA-seq data using causal generative adversarial networks. Nat Commun [Epub ahead of print]. [article]