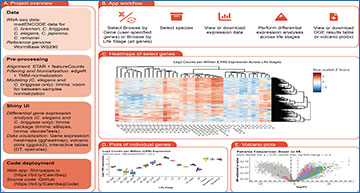
The Caenorhabditis RNA-seq Browser is an open-source Shiny web app that enables on-demand visualization and quantification of bulk RNA-sequencing data for five Caenorhabditis species: C. elegans, C. briggsae, C. brenneri, C. japonica, and C. remanei. The app is designed to allow researchers without previous coding experience to interactively explore publicly available Caenorhabditis RNA-sequencing data. Key app features include the ability to plot gene expression across life stages for user-specified gene sets, and modules for performing differential gene expression analyses.
Diagram of the Caenorhabditis RNA-seq Browser workflow
and examples of the Shiny user interface
A) Overview of this project, including the data included, our pre-processing workflow, features of the Shiny user interface, and deployment information. B) User workflow for the Caenorhabditis RNA-seq Browser. C) Heatmap generated by the app in “Browse by Gene” mode. Search term used to generate the image was “GPCR”; species is C. elegans. Heatmap rows reflect log2CPM values of individual genes and are ordered using Pearson clustering; columns reflect individual samples (life stages) are ordered by Spearman clustering. D) Plots of log2CPM values for an individual gene (C. elegans npr-1) across life stages. E) Volcano plot of differential gene expression comparing C. elegans dauers and young adults.
Availability – The Caenorhabditis RNA-seq Browser can be accessed online via shinyapps.io or can be installed locally in R from a GitHub repository.
Akimori D, Hillier LW, Bryant AS. (2024) The Caenorhabditis RNA-seq Browser: a web-based application for on-demand analysis of publicly available Caenorhabditis spp. bulk RNA-sequencing data. microPublication [Epub ahead of print]. [article]