Bulk RNA-Seq remains a widely adopted technique to profile gene expression, primarily due to the persistent challenges associated with achieving single-cell resolution. However, a key challenge is accurately estimating the proportions of different cell types within these bulk samples. To address this issue, University of Helsinki researchers have developed DeconV, a probabilistic framework for cell-type deconvolution that uses scRNA-Seq data as a reference. This approach aims to mitigate some of the limitations in existing methods by incorporating statistical frameworks developed for scRNA-Seq, thereby simplifying issues related to reference preprocessing such as normalization and marker gene selection. The researchers benchmarked DeconV against established methods, including MuSiC, CIBERSORTx, and Scaden. Their results show that DeconV performs comparably in terms of accuracy to the best-performing method, Scaden, but provides additional interpretability by offering confidence intervals for its predictions. Furthermore, the modular design of DeconV allows for the investigation of discrepancies between bulk-sequenced samples and artificially generated pseudo-bulk samples.
Overview of DeconV methodology
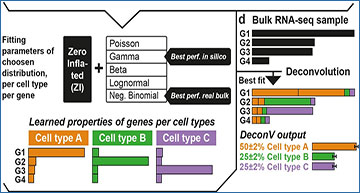
a. scRNA-seq example of a reference dataset, containing three cell types. Three marker and one non informative genes are represented in the heatmap. b. The probabilistic framework of DeconV leads to an implicit weighing of genes during deconvolution, as the low dispersion of marker genes in their cell type will have a stronger effect during the fitting phase than a non informative gene. c. Parameters of the chosen distribution are then learned from the reference dataset. Benchmarks show that zero-inflated Gamma distribution is performing best with in silico bulk RNA-seq reconstructed from scRNA-seq but zero-inflated negative binomial has better results with real bulk RNA-Seq. d. Cell-types proportions are estimated from the query dataset after maximizing the likelihood of genes and computing the best fit. In this schematic, there is two times more cell type A than B or C. DeconV also provides the confidence intervals (default at 95%) of the estimated proportion, in this example 2% for each cell type.
Gynter A, Meistermann D, Lähdesmäki H, Kilpinen H. (2023) DeconV: Probabilistic Cell Type Deconvolution from Bulk RNA-sequencing Data. bioRXiv [online pre-print]. [article]