Wistar scientists have developed a new tool that can help identify cancer-associated microbes by using machine learning technology. Under the leadership of Dr. Noam Auslander — assistant professor in the Ellen and Ronald Caplan Cancer Center’s Molecular & Cellular Oncogenesis Program — the group has analyzed short read RNA-sequencing data to detect biomarkers for esophageal carcinoma, or ESCA. Their paper, “Microbial gene expression analysis of healthy and cancerous esophagus uncovers bacterial biomarkers of clinical outcomes,” was published in International Society for Microbial Ecology Communications.
Tumor microenvironments are often analyzed using RNA sequencing, or RNAseq, which identifies mRNA in a population of cells to find which genes are being expressed. Theoretically, RNAseq data can reveal the expression of microbial genes in cancerous tissue, which could help to identify microbiome shifts that might be playing a role in the cancer’s development. But RNAseq “reads” — the physical lengths of genetic data that correspond with gene expression — are often quite short, posing a challenge for classifying them into diverse microbial genetic origins. Assembling the short RNAseq reads into longer contiguous segments that can be associated with a vast array of potential origins — be they human, viral, or bacterial — to identify specific microbes whose expression correlates with ESCA is computationally challenging.
That’s where Dr. Auslander and her group decided to intervene by training a convolutional neural network, a type of machine-learning technology that can be taught to train itself to accurately assess large quantities of data. The team, using large publicly available datasets of characterized short-read data, trained the network to sort short-read RNAseq data by its likely origin: human, viral, or bacterial. Their model sought to pare down the number of short reads that would need to be assembled for identification, which would reduce the computational load of screening for microbial influences in cancer tissue.
Once the model was trained, its sorting capabilities allowed the group to selectively analyze ESCA tissue for reads of microbial origin and compare those data with apparently healthy esophageal tissue. Auslander’s team found several instances of microbial expression present in ESCA with significantly less incidence in apparently healthy esophageal tissue.
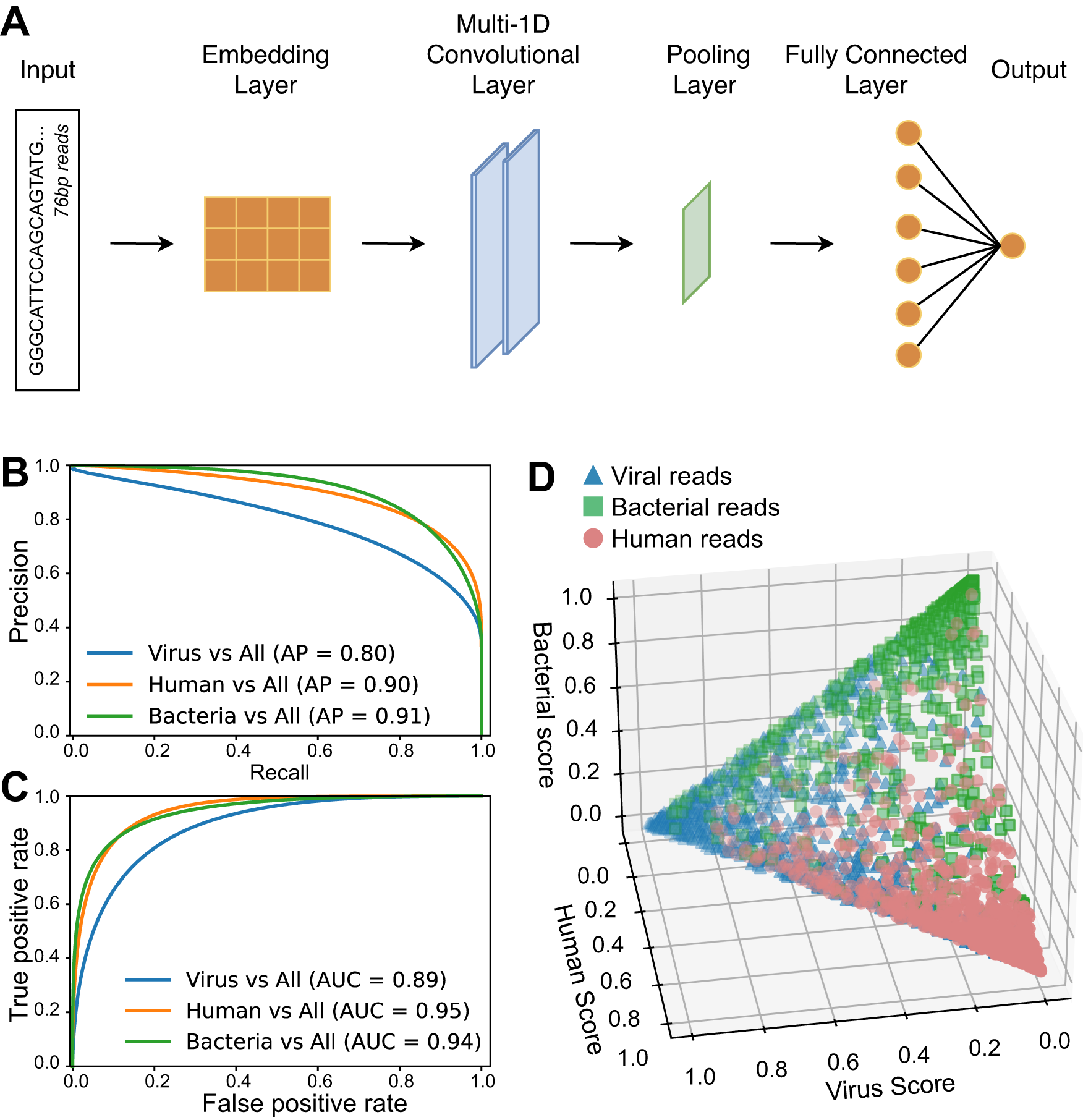
Read-classification model architecture and performance
A Overview of the model architecture. B Test-set one-versus-all precision-recall curves for each class of sequence origin. C Test-set one-versus-all receiver-operating characteristic curves for each class. The AUCs are the areas under each curve. D Model scores for 1000 randomly-selected sequences from each class, plotted on the x + y + z = 1 plane.
In particular, they found that nearly half of the microbial genes over-expressed in cancer originated from bacteriophages, which are viruses that infect bacteria; this finding may indicate that viruses infecting microorganisms within the tumor microenvironment facilitate ongoing cancerous gene expression.
The team also identified patient outcome predictors amid the data. Bacterial iron-sulfur proteins were found to impact human genes involved in ferroptosis — a type of cell death pathway that’s modulated by iron — which predicted poor prognosis in ESCA patients. Microbial genes involved in mitochondrial reprogramming were also found to predict ESCA patient prognosis.
“By building on our previous work, our team has successfully leveraged machine learning to dive deeper into what’s going on inside cancer,” said Dr. Auslander. “While it’s always important to remember that correlation does not equal causation, the associations we’ve been able to find between certain microbial genes and ESCA will allow scientists to further understand the mechanics of esophageal cancer — which is the first step in stopping it.”
Source – The Wistar Institute
Availability – viRNAtrap-bacteria is available at: https://github.com/AuslanderNoam/virnatrap-bacteria
Schäffer DE, Li W, Elbasir A, Altieri DC, Long Q, Auslander N. (2023) Microbial gene expression analysis of healthy and cancerous esophagus uncovers bacterial biomarkers of clinical outcomes. ISME Commun 3(1):128. [article]