How can we study RNA that is in the process of being synthesized? For over half a century, researchers have developed diverse approaches to isolate and sequence nascent RNA and reveal layers of transcriptional control that are invisible when capturing only mature RNA. For example, nascent RNA sequencing (RNA-seq) can provide insight into polymerase movement, transcription rates, and unstable or noncoding RNAs.
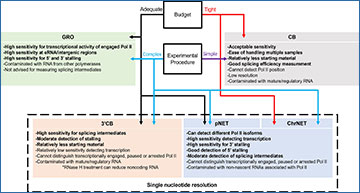
Historically, there are three major classes of nascent RNA-seq protocols. First, there are approaches that rely on nuclear run-on assays, such as global run-on-sequencing (GRO-seq). During the run-on assay, a nucleotide analog is incorporated into actively transcribed RNA to allow immunoprecipitation of nascent RNA. Second, nascent RNA can be captured by immunoprecipitating RNA polymerase II (Pol II), such as in plant native elongating transcript sequencing (pNET-seq). Finally, after chromatin isolation, Pol II can remain bound and chromatin-bound (CB) RNA can be sequenced (Weber et al. 2014). Each of these methods has strengths and weaknesses, making it difficult to choose the best approach for specific research needs.
In the latest issue of The Plant Cell, Min Liu and colleagues systematically compare the major nascent RNA-seq approaches—GRO-seq, pNET-seq, and CB RNA-seq—and provide detailed guidance for selecting a method.
Liu M, Zhu J, Huang H, Chen Y, Dong Z. (2023) Comparative analysis of nascent RNA sequencing methods and their applications in studies of cotranscriptional splicing dynamics. Plant Cell 35(12):4304-4324. [abstract]