The 10x Genomics Chromium single-cell RNA sequencing technology is a powerful gene expression profiling platform, which is capable of profiling expression of thousands of genes in tens of thousands of cells simultaneously. This platform can produce hundreds of million reads in a single experiment, making it a very challenging task to quantify expression of genes in individual cells due to the massive data volume. Researchers at La Trobe University have developed cellCounts, a new tool for efficient and accurate quantification of Chromium data. cellCounts employs the seed-and-vote strategy to align reads to a reference genome, collapses reads to UMIs (Unique Molecular Identifiers) and then assigns UMIs to genes based on the featureCounts program. Using both simulation and real datasets for evaluation, cellCounts was found to compare favorably to cellRanger and STARsolo. cellCounts is implemented in R, making it easily integrated with other R programs for analysing Chromium data.
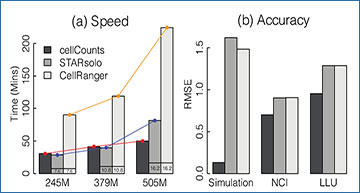
Comparison of speed and accuracy for quantifying 10x Chromium scRNA-seq data
(a) Running time of cellCounts, STARsolo and CellRanger on three real datasets. Number of reads included in each dataset is indicated under each column. These datasets all have a BCL format. Numeric values shown at the bottom of bars for STARsolo and CellRanger indicate the amount of time spent on converting BCL-format reads to FASTQformat reads. cellCounts’s running time does not include this time because cellCounts directly processes BCL-format reads. (b) RMSE (root mean square error) of gene expression calculated for cellCounts, STARsolo and CellRanger based on ground truth. For each method, UMI counts were converted to log2-cpm values for each gene in each cell and then used for calculating RMSE. An offset of 0.5 was added to UMI counts to avoid log transformation of zero counts. ‘Simulation’ is a simulation dataset. ‘NCI’ and ‘LLU’ are two real datasets generated from sequencing of two human cell lines mixed at known ratios.
Availability – cellCounts was implemented as a function in R package Rsubread that can be downloaded from http://bioconductor.org/packages/release/bioc/html/Rsubread.html.
Liao Y, Raghu D, Pal B, Mielke LA, Shi W. (2023) cellCounts: an R function for quantifying 10x Chromium single-cell RNA sequencing data. Bioinformatics [Epub ahead of print]. [abstract]