The development of single-cell RNA sequencing (scRNA-seq) technology has enabled researchers to study gene expression at the single-cell level, providing insights into cellular heterogeneity and dynamic cellular processes. However, most scRNA-seq methods focus on static snapshots of gene expression, providing limited information about temporal changes in gene expression over time. Researchers from Xiamen University introduce a new scRNA-seq method called Well-TEMP-seq, which allows for the massively parallel profiling of single-cell temporal RNA dynamics.
The Well-TEMP-seq method involves capturing single cells in microwells and inducing RNA labeling with a temperature-sensitive RNA labeling reagent. Cells are then exposed to a series of temperature steps, which activate RNA labeling at different time points. After RNA labeling, cells are pooled and subjected to scRNA-seq analysis, allowing for the simultaneous profiling of single-cell temporal RNA dynamics.
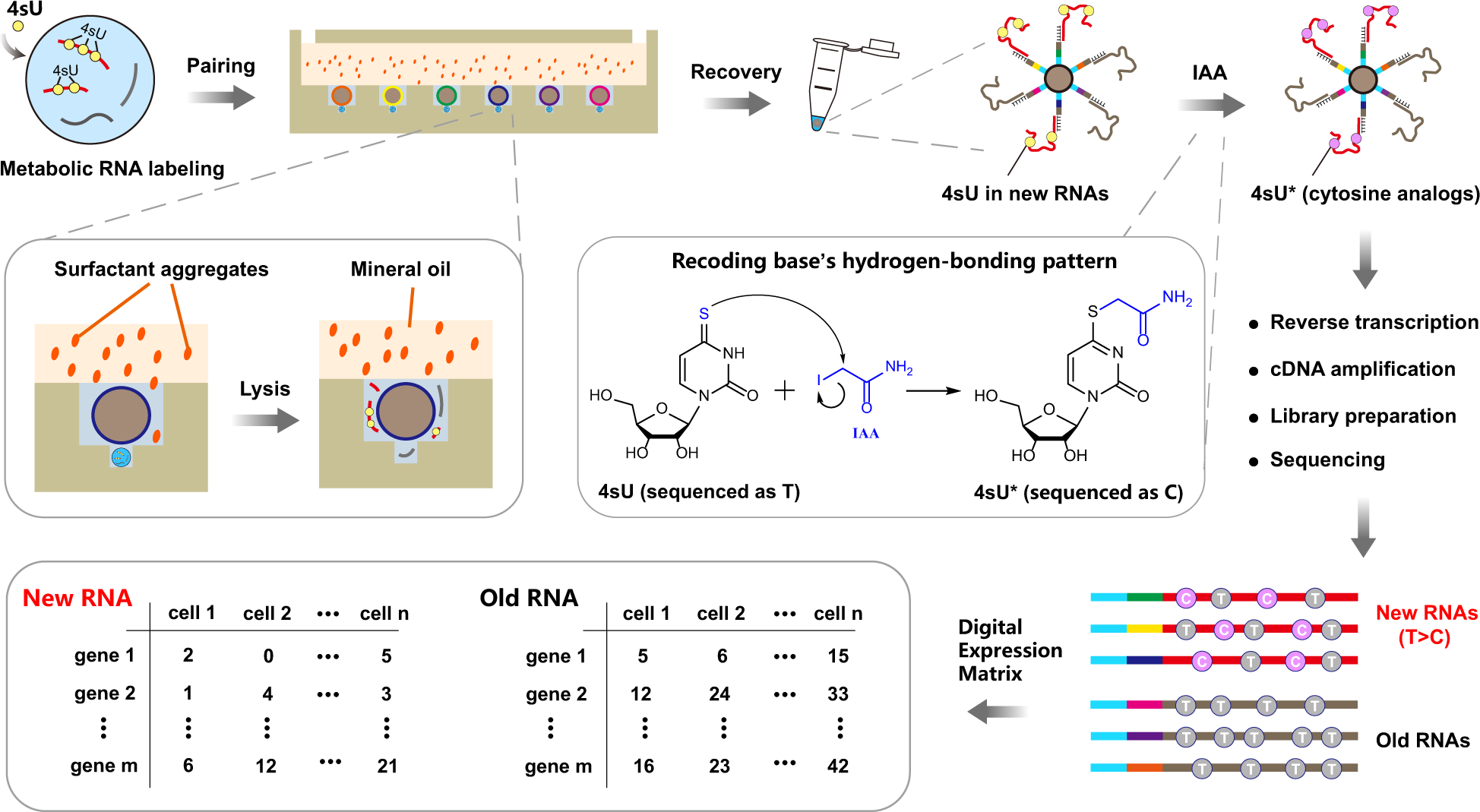
Overview and workflow of Well-TEMP-seq
Cells are first incubated with 4sU to label newly transcribed RNAs and are then loaded into Well-paired-seq chip for single cell/bead pairing. After cell lysis by surfactant aggregates (sakosyl), RNAs are captured by DNA-barcoded beads and subjected to IAA (iodoacetamide) treatment to transform 4sU to a cytosine analog (4sU*), which induces T-to-C substitutions at 4sU-labeled sites. RNAs on the beads are reverse transcribed and the resulting cDNAs are amplified for library preparation and sequencing. Reads with T-to-C substitution(s) are identified as new RNAs while others are old RNAs. Therefore, each cell is associated with two digital expression matrices (new RNA and old RNA).
The researchers demonstrated the effectiveness of Well-TEMP-seq in profiling temporal gene expression changes in single cells. They used Well-TEMP-seq to study the differentiation of mouse embryonic stem cells (mESCs) into definitive endoderm (DE) cells, a process characterized by dynamic changes in gene expression over time. The researchers compared the results of Well-TEMP-seq to those of a conventional scRNA-seq method and found that Well-TEMP-seq enabled the identification of DE-specific gene expression dynamics that were not captured by conventional scRNA-seq.
The researchers also used Well-TEMP-seq to study the response of human mammary epithelial cells (HMECs) to treatment with the drug tamoxifen. Tamoxifen is commonly used to treat breast cancer, and its response is characterized by temporal changes in gene expression. Using Well-TEMP-seq, the researchers were able to identify different clusters of cells with distinct temporal gene expression patterns in response to tamoxifen treatment.
Overall, the results of the study demonstrate the effectiveness of Well-TEMP-seq in capturing temporal changes in gene expression at the single-cell level. The method has the potential to provide new insights into dynamic cellular processes, such as cell differentiation and drug response, that cannot be captured by conventional scRNA-seq methods.
In conclusion, these researchers introduce a new scRNA-seq method that enables the simultaneous profiling of single-cell temporal RNA dynamics. The results of the study demonstrate the effectiveness of the method in capturing temporal changes in gene expression and its potential for providing new insights into dynamic cellular processes. The development of Well-TEMP-seq represents a significant advancement in scRNA-seq technology and has the potential to drive new discoveries in the field of single-cell genomics.
Lin S, Yin K, Zhang Y, Lin F, Chen X, Zeng X, Guo X, Zhang H, Song J, Yang C. (2023) Well-TEMP-seq as a microwell-based strategy for massively parallel profiling of single-cell temporal RNA dynamics. Nat Commun 14(1):1272. [article]