Behavior arises from concerted activity throughout the brain. Consequently, a major focus of modern neuroscience is defining the physiology and behavioral roles of projection neurons linking different brain areas. Single-cell RNA sequencing has facilitated these efforts by revealing molecular determinants of cellular physiology and markers that enable genetically targeted perturbations such as optogenetics, but existing methods for sequencing defined projection populations are low throughput, painstaking, and costly.
Researchers at the University of California, San Francisco have developed a straightforward, multiplexed approach, virally encoded connectivity transgenic overlay RNA sequencing (VECTORseq). VECTORseq repurposes commercial retrogradely infecting viruses typically used to express functional transgenes (e.g., recombinases and fluorescent proteins) by treating viral transgene mRNA as barcodes within single-cell datasets. VECTORseq is compatible with different viral families, resolves multiple populations with different projection targets in one sequencing run, and identifies cortical and subcortical excitatory and inhibitory projection populations. This study provides a roadmap for high-throughput identification of neuronal subtypes based on connectivity.
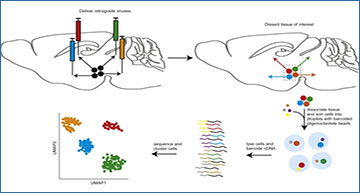
Schematic of VECTORseq

Injecting retrogradely infecting viruses (colored syringes) into brain structures (sagittal section of mouse brain in this schematic) targeted by different projection neurons (directed arrows) from a single structure of origin will label each with unique virally encoded RNA barcodes. Following standard single-cell sequencing methods and analysis, the expression of viral barcodes can be overlaid to assign each cluster to its projection target.
Cheung V, Chung P, Bjorni M, Shvareva VA, Lopez YC, Feinberg EH. (2022) Virally encoded connectivity transgenic overlay RNA sequencing (VECTORseq) defines projection neurons involved in sensorimotor integration. Cell Rep 37(12):110131. [article]