Acute myeloid leukemia (AML) is a type of cancer that affects the bone marrow and blood. It is a very aggressive cancer and has a high mortality rate. One of the latest advances in cancer research is the use of CAR-T cell therapy. This therapy uses a patient’s own T cells, which are genetically modified to recognize and attack cancer cells.
However, CAR-T cell therapy can be challenging to develop because not all patients respond to it, and the development of CAR-T cells can be a long and costly process. Recently, researchers from the University Hospital, LMU Munich used single-cell RNA sequencing (scRNA-seq) to help guide the development of CAR-T cells for the treatment of AML.
ScRNA-seq is a technique that allows researchers to study the transcriptomes of individual cells. This technique has revolutionized the field of genomics, allowing researchers to study gene expression in individual cells and identify previously unknown cell types. In this publication, the researchers used scRNA-seq to identify the cell types present in AML samples and to guide the development of CAR-T cells.
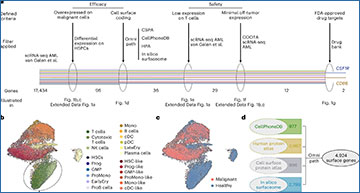
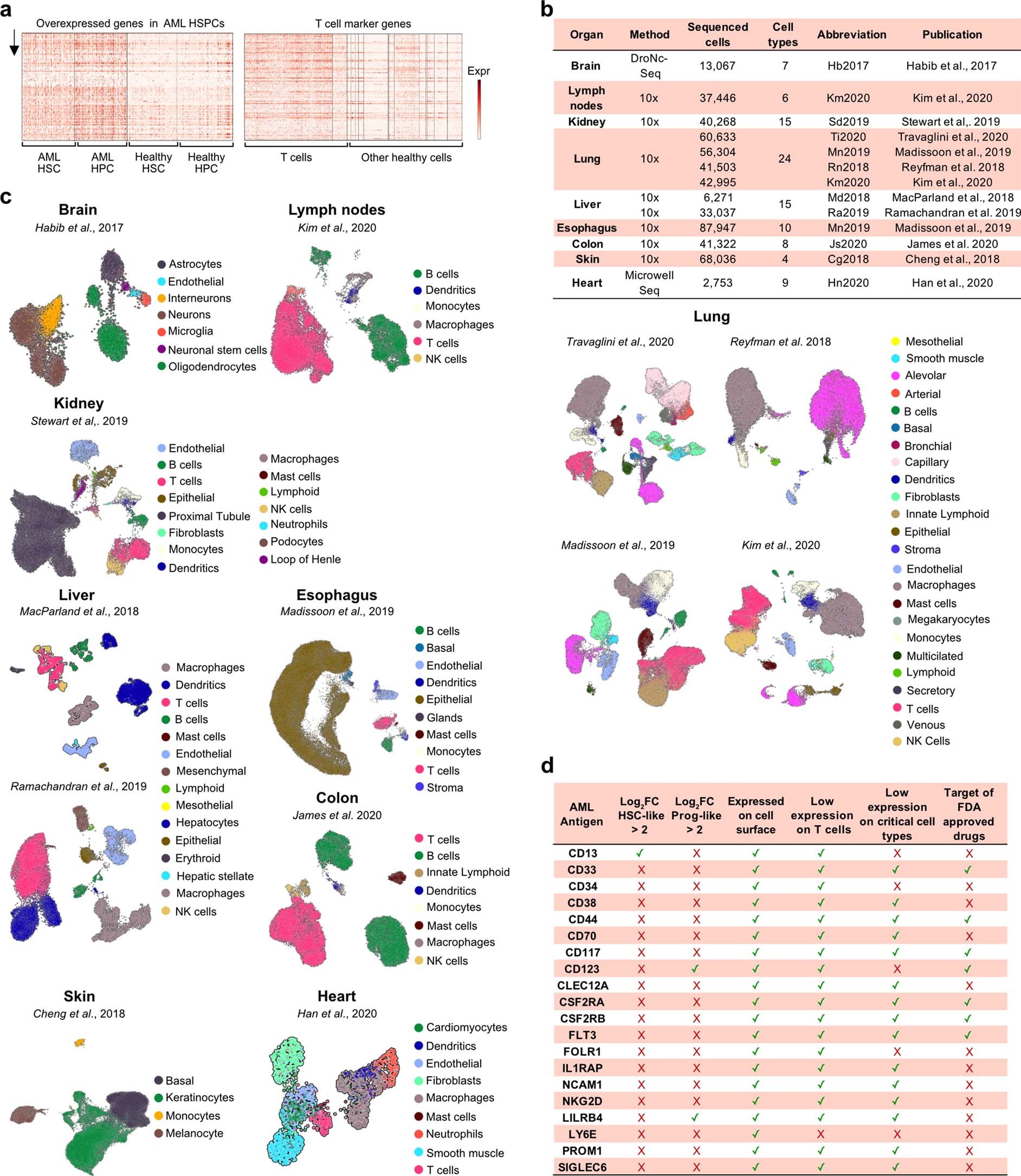
The researchers leveraged an atlas of publicly available RNA-sequencing data of over 500,000 single cells from 15 individuals with AML and tissue from 9 healthy individuals for prediction of target antigens that are expressed on malignant cells but lacking on healthy cells, including T cells. They then used this information to design CAR-T cells that would target the cancer cells specifically.
Summary of the cross-organ off-target transcriptomic atlas (COOTA)
(a) Top 100 overexpressed genes in AML HSPC (left) and healthy T cells (right) from differential expression analysis. Normalized expression values were logarithmized and scaled to unit variance. (b) Overview of 11 scRNA-seq datasets of various healthy tissues used to quantify off-target antigen expression. (c) UMAP plots of 11 scRNA-seq datasets with colors highlighting clustering into respective cell types. Cell annotations were provided by the authors of the respective studies. (d) Current CAR targets in AML were cross-referenced to filters used for the single cell-based target screening approach. OE HSC-/Prog-like: overexpressed on HSC-/Prog-like cells with log fold change > 2 and FDR-adjusted p ≤ 0.01, using a t-test with overestimated variance. Red cross: Antigen did not fulfill the respective threshold or criteria. Green check: Thresholds or criteria were passed.
The researchers also tested the CAR-T cells in vitro and in vivo. They found that the CAR-T cells were able to specifically target the cancer cells and effectively kill them. They also found that the CAR-T cells were able to reduce tumor burden and prolong the survival of mice with AML.
This study demonstrates the power of scRNA-seq in guiding the development of CAR-T cell therapy. By identifying the cell types present in AML and designing CAR-T cells that specifically target the cancer cells, the researchers were able to develop a more effective and efficient therapy. This approach could be applied to other types of cancer and could help accelerate the development of CAR-T cell therapies in the future.
Gottschlich A, Thomas M, Grünmeier R, Lesch S, Rohrbacher L, Igl V, Briukhovetska D, Benmebarek MR, Vick B, Dede S, Müller K, Xu T, Dhoqina D, Märkl F, Robinson S, Sendelhofert A, Schulz H, Umut Ö, Kavaka V, Tsiverioti CA, Carlini E, Nandi S, Strzalkowski T, Lorenzini T, Stock S, Müller PJ, Dörr J, Seifert M, Cadilha BL, Brabenec R, Röder N, Rataj F, Nüesch M, Modemann F, Wellbrock J, Fiedler W, Kellner C, Beltrán E, Herold T, Paquet D, Jeremias I, von Baumgarten L, Endres S, Subklewe M, Marr C, Kobold S. (2023) Single-cell transcriptomic atlas-guided development of CAR-T cells for the treatment of acute myeloid leukemia. Nat Biotechnol [Epub ahead of print]. [abstract]