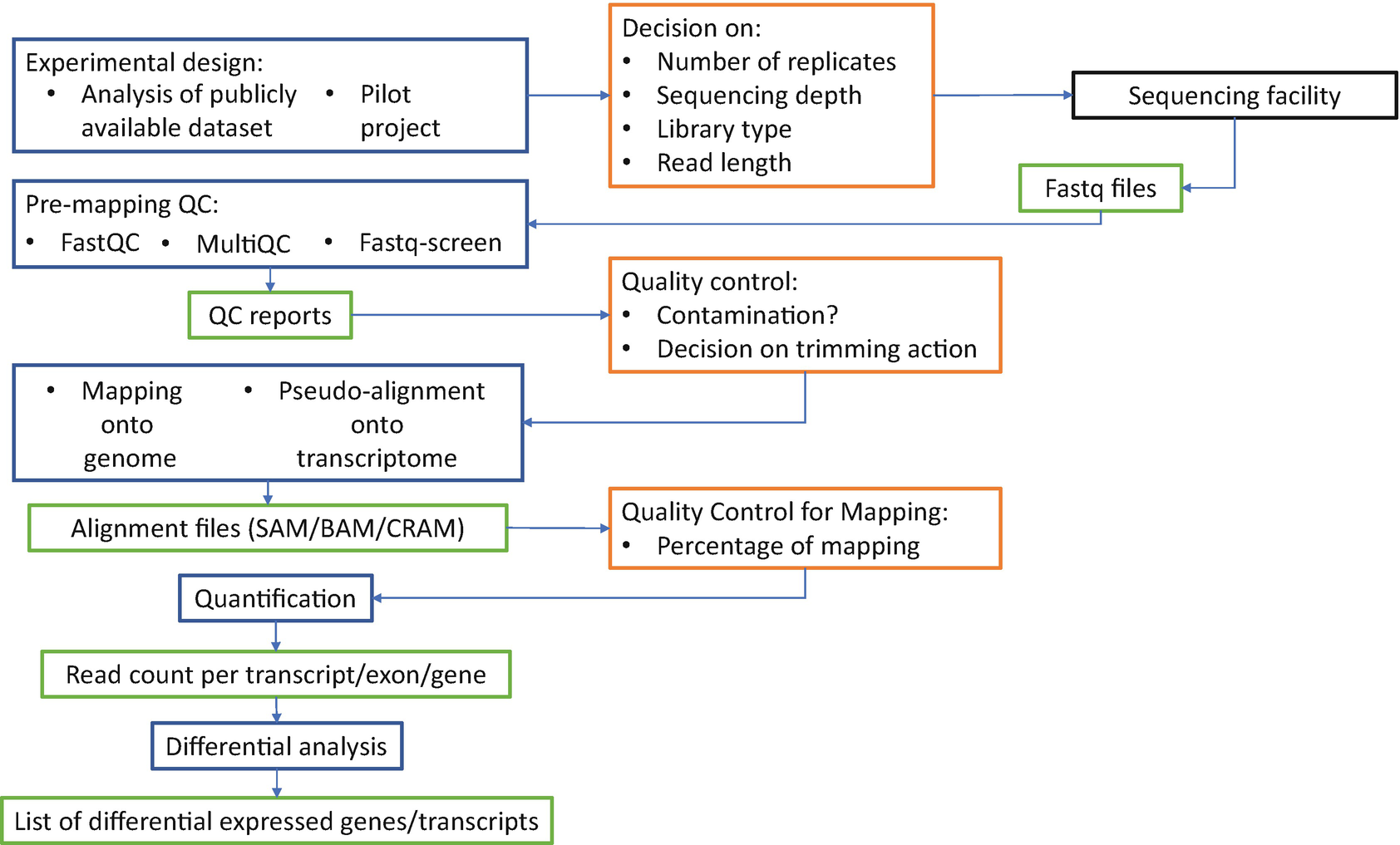
Ribonucleic acids (RNAs) are fundamental molecules that control regulation and expression of the genome and therefore the function of a cell. Robust analysis and quantification of RNA transcripts hold critical importance in understanding cell function, altered phenotypes in different biological context, for understanding and targeting diseases. The development of RNA-sequencing (RNA-Seq) now provides opportunities to analyze the expression and function of RNA molecules at an unprecedented scale. However, the strategy for RNA-Seq experimental design and data analysis can substantially differ depending on the biological application. The design choice could also have significant impact for downstream results and interpretation of data.
Researchers from the University of Otago describe key critical considerations required for RNA-Seq experimental design and also describe a step-by-step bioinformatics workflow detailing the different steps required for RNA-Seq data analysis. The authors believe this article will be a valuable guide for designing and analyzing RNA-Seq data to address a wide range of different biological questions.
Gimenez G, Stockwell PA, Rodger EJ, Chatterjee A. (2023) Strategy for RNA-Seq Experimental Design and Data Analysis. Methods Mol Biol 2588:249-278. [abstract]