Single cell RNA sequencing has the potential to elucidate transcriptional programs underlying key cellular phenotypes and behaviors. However, many cell phenotypes are incompatible with indiscriminate single cell sequencing because they are rare, transient, or can only be identified by imaging. Existing methods for isolating cells based on imaging for single cell sequencing are technically challenging, time-consuming, and prone to loss because of the need to physically transport single cells.
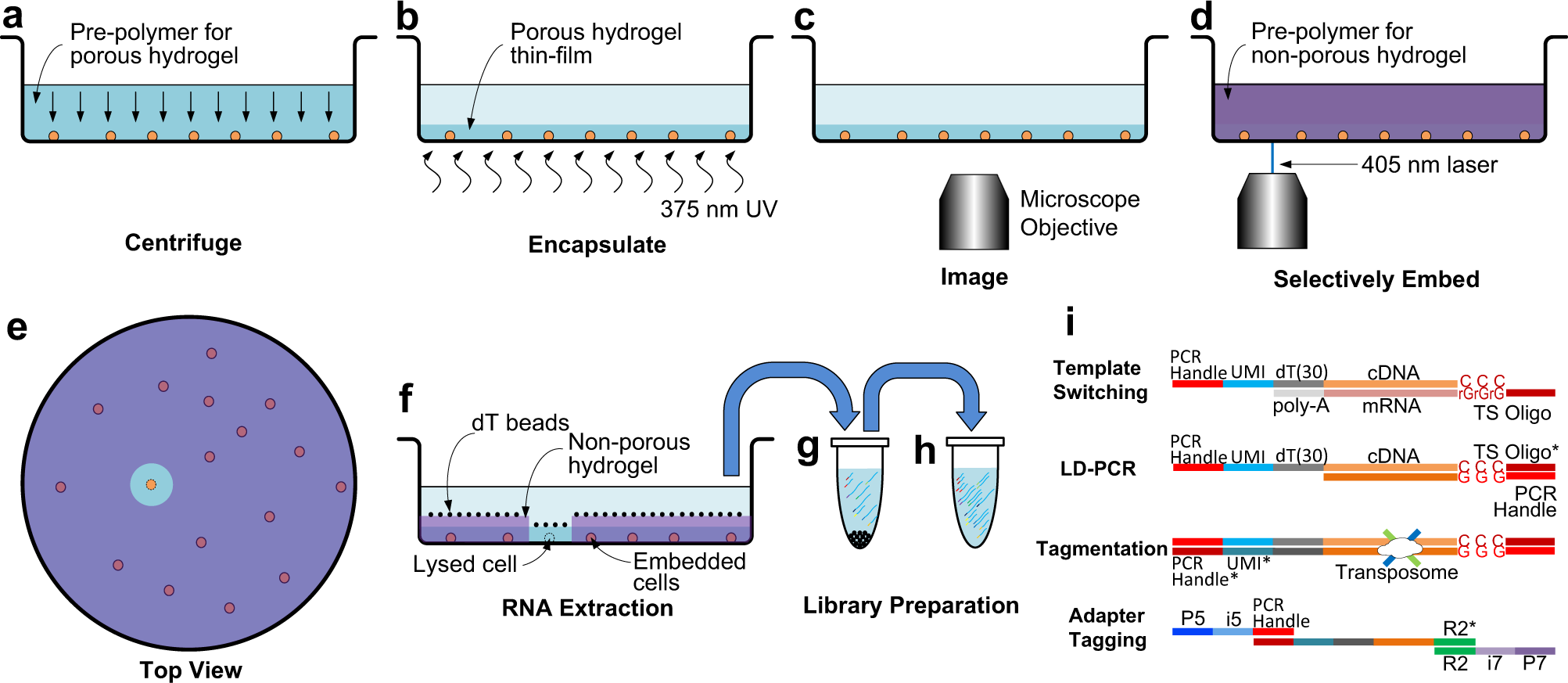
Researchers at the University of British Columbia have developed See-N-Seq, a method to rapidly screen cells in microwell plates in order to isolate RNA from specific single cells without needing to physically extract each cell. This approach involves encapsulating the cell sample in a micropatterned hydrogel with spatially varying porosity to selectively expose specific cells for targeted RNA extraction. Extracted RNA can then be captured, barcoded, reverse transcribed, amplified, and sequenced at high-depth. The researchers used See-N-Seq to isolate and sequence RNA from cell-cell conjugates forming an immunological synapse between T-cells and antigen presenting cells. In the hours after synapsing, they found time-dependent bifurcation of single cell transcriptomic profiles towards Type 1 and Type 2 helper T-cells lineages. These results demonstrate how See-N-Seq can be used to associate transcriptomic data with specific functions and behaviors in single cells.
Selective single cell RNA sequencing using See-N-Seq
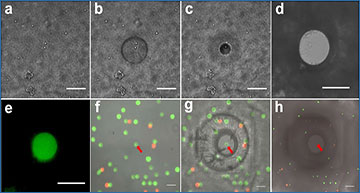
a The cell sample is distributed in imaging microwell plates with the pre-polymer for the porous hydrogel and aligned by centrifuging. b The cells are encapsulated in a porous hydrogel thin-film formed by exposure to 375 nm UV light. c Encapsulated cells are imaged by microscopy to identify a target cell in each well. d The pre-polymer for the non-porous hydrogel penetrates inside the porous hydrogel and is micropatterned using a scanning 405 nm laser. e (Top view) All cells except for the target cell are embedded in the non-porous hydrogel. f mRNA from the target cell is captured by oligo-dT beads. g The oligo-dT beads are transferred to a tube where mRNA is eluted, UMI-barcoded, and reverse transcribed with a template switching oligo. h The cDNA library is amplified by long distance PCR (LD-PCR). i Library preparation steps include template switching, LD-PCR, tagmentation, and adaptor tagging.
Lee JH, Park ES, Choi JR, Matthews K, Lam AV, Deng X, Duffy SP, Ma H. (2022) See-N-Seq: RNA sequencing of target single cells identified by microscopy via micropatterning of hydrogel porosity. Commun Biol 5(1):768. [article]