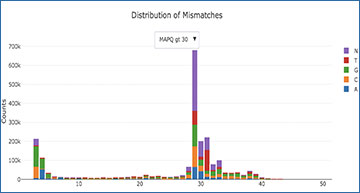
SAMStat is an efficient program to extract quality control metrics from fastq and SAM/BAM files. A distinguishing feature is that it displays sequence composition, base quality composition and mapping error profiles split by mapping quality. This allows users to rapidly identify reasons for poor mapping including the presence of untrimmed adapters or poor sequencing quality at individual read positions.
Researchers from the University of Western Australia present a major update to SAMStat. The new version now supports paired-end and long read data. Quality control plots are drawn using the ploty javascript library.
The output is a single html page. SAMStat reports length distribution, base quality distribution, mapping statistics, mismatch, insertion and deletion error profiles.
Availability – The source code of SAMStat and code to reproduce the results are found here: https://github.com/timolassmann/samstat.
Lassmann T. (2023) SAMStat 2: quality control for next generation sequencing data. Bioinformatics [Epub ahead of print]. [abstract]