With development of many effective approaches to fighting cancer, the focus becomes less about finding new ways to treat patients and more about identifying which already available treatments a patient will respond best to. Immune checkpoint inhibitors have emerged as an effective way to boost the cancer-fighting ability of T cells. Unsurprisingly, some patients are much more responsive to immune checkpoint inhibitors than others. In kidney or renal cancer, such as renal cell carcinoma (RCC), the types of T cells present in the tumor at the time of diagnosis often correlate with a patient’s tumor responsiveness to immune checkpoint inhibitor treatment.
The problem is that “most of the biomarker studies have uniformly analyzed tumor-infiltrating lymphocyte (TIL) populations in total and resulting biomarker associations with clinical outcomes remain imprecise,” explains postdoctoral researcher Dr. Yuexin Xu, from the lab of Dr. Edus ‘Hootie’ Warren, Professor in the Clinical Research and Vaccine and Infectious Disease Divisions and Head of the Global Oncology Program at Fred Hutch. In their recent Frontiers in Oncology study, the researchers thought “single-cell gene expression profiling of individual tumor-infiltrating T cells from clear cell RCC [was] necessary to decode the function of T cells at the clonal level,” Dr. Xu notes.
To investigate the spatial heterogeneity of infiltrating T cells in renal tumors and to understand how they clonally expand, the authors collected renal tumors and normal adjacent tissue, in addition to blood cells (PBMCs) from 14 patients. These RCC patients consisted of two subtypes of renal cancer- clear cell and non-clear cell RCC which are known to respond differently to treatments. Xu et al. identified distinct patterns of tumor T cell infiltration between the different tumor subtypes, with significantly higher infiltration in clear cell RCC compared to non-clear cell RCC tumors. T cell receptor diversity is indicative of a variety of different foreign antigens that T cells recognize. When T cells encounter an antigen that matches their specific receptor, they then clonally expand to boost their ability to fight off these foreign cells. The authors then investigated T cell receptor diversity and clonal composition of T cells infiltrating into renal tumors. Interestingly, expanded T cell clones in clear cell RCC carry T cell receptors that are not commonly shared with healthy subjects, suggesting that the clonally expanded T cells may be reacting against RCC-associated targets. Furthermore, the authors found that RCC tumor infiltrating T cell repertoires were unique to individuals, rather than sharing common antigen targets across RCC tumors.
T cell expansion in renal cell carcinoma.Image provided by Yuexin Xu. Created with BioRender
The authors then turned to single cell gene expression methods to investigate the phenotypes and functional capacity of clonally expanded tumor infiltrating T cells. Again, isolating T cells from renal tumors in addition to normal adjacent tissue and blood, Xu et al. then performed single cell RNA sequencing (scRNA-seq).
Dr. Xu exclaims, that this approach “enabled us to study the same T cell clone’s phenotype in different locations, normal and tumor. Learning about the tumor-infiltrating T cell repertoire structures and tracking the functional phenotype of individual T cell clones fascinates me. To our great surprise, many of the tumor-infiltrating effector T cell clones that [were]shared with the PBMC [samples]retain their functional phenotype.”
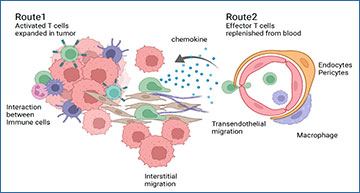
The authors found that expanded T cell clones shared across all tissues consisted of effector T cells that expressed genes indicative of T cell migration. In contrast, T cell clones specific to tumor samples were enriched for the memory T cell lineage and had increased expression of genes indicative of T cell exhaustion, those involved in MHC antigen presentation, and signaling marks consistent with target antigen recognition in the tumor microenvironment. Furthermore, these tumor-restricted T cells had increased expression of the proliferation marker, MKI67. These findings suggest that expanded T cell clones remained capable of continued proliferation and expansion within the tumor microenvironment. The authors confirmed this with immunohistochemistry and found that proliferating T cells were indeed located in close proximity to tumor cells, supporting direct interactions between expanding T cell populations and tumor cells.
Xu notes, “We think this work established a framework to streamline single-cell RNA-seq analysis of TIL, T-cell repertoires analysis, and multiplex IHC to study the heterogeneous RCC tumor microenvironment for the individual patient. Knowing the great heterogeneity of tumoral structure and immune-infiltrating patterns among ccRCC patients, this framework could be potentially applied to the current TIL therapy, to survey the TCR repertoire and potentially guild the selection of TIL for therapy for an individual.”
Dr. Xu adds, “We discovered two different expanding tumor-infiltrating T-cell populations in ccRCC, with distinct phenotypes: a tumor-restricted T-cell population with exhausted, memory-like phenotype and a shared T-cell population shared with blood with an effector phenotype, expressing higher cytotoxic markers but lower exhaustion and activation markers than the tumor-restricted ones. The expression of chemokine and chemokine receptors of these two populations are different, suggesting two distinct homing and activation routes of these T-cell clones.” Moving forward, the authors plan to “use CRISPR-Cas9 screening technology to identify the potential [ccRCC] antigen targets” in addition to “exploring the option of spatial genomics, to illustrate the potential interactions of tumor-infiltrating T cells with the microenvironment,” Xu explains.
Dr. Xu recognizes Dr. Scott Tykodi and her mentor, Dr. Edus Warren, for their support and guidance on this project, in addition to Dr. Shreeram Akilesh from UW Pathology who helped with analysis of patient IHC data.
Source – Fred Hutchinson Cancer Center
Xu Y, Morales AJ, Towlerton AMH, Akilesh S, Miller CP, Tykodi SS, Warren EH. (2022) Integrated TCR repertoire analysis and single-cell transcriptomic profiling of tumor-infiltrating T cells in renal cell carcinoma identifies shared and tumor-restricted expanded clones with unique phenotypes. Front Oncol 12:952252. [article]