Spatial transcriptomics (ST) measures mRNA expression across thousands of spots from a tissue slice while recording the two-dimensional (2D) coordinates of each spot. Princeton University researchers have developed probabilistic alignment of ST experiments (PASTE), a method to align and integrate ST data from multiple adjacent tissue slices. PASTE computes pairwise alignments of slices using an optimal transport formulation that models both transcriptional similarity and physical distances between spots. PASTE further combines pairwise alignments to construct a stacked 3D alignment of a tissue. Alternatively, PASTE can integrate multiple ST slices into a single consensus slice. The researchers show that PASTE accurately aligns spots across adjacent slices in both simulated and real ST data, demonstrating the advantages of using both transcriptional similarity and spatial information. They further show that the PASTE integrated slice improves the identification of cell types and differentially expressed genes compared with existing approaches that either analyze single ST slices or ignore spatial information.
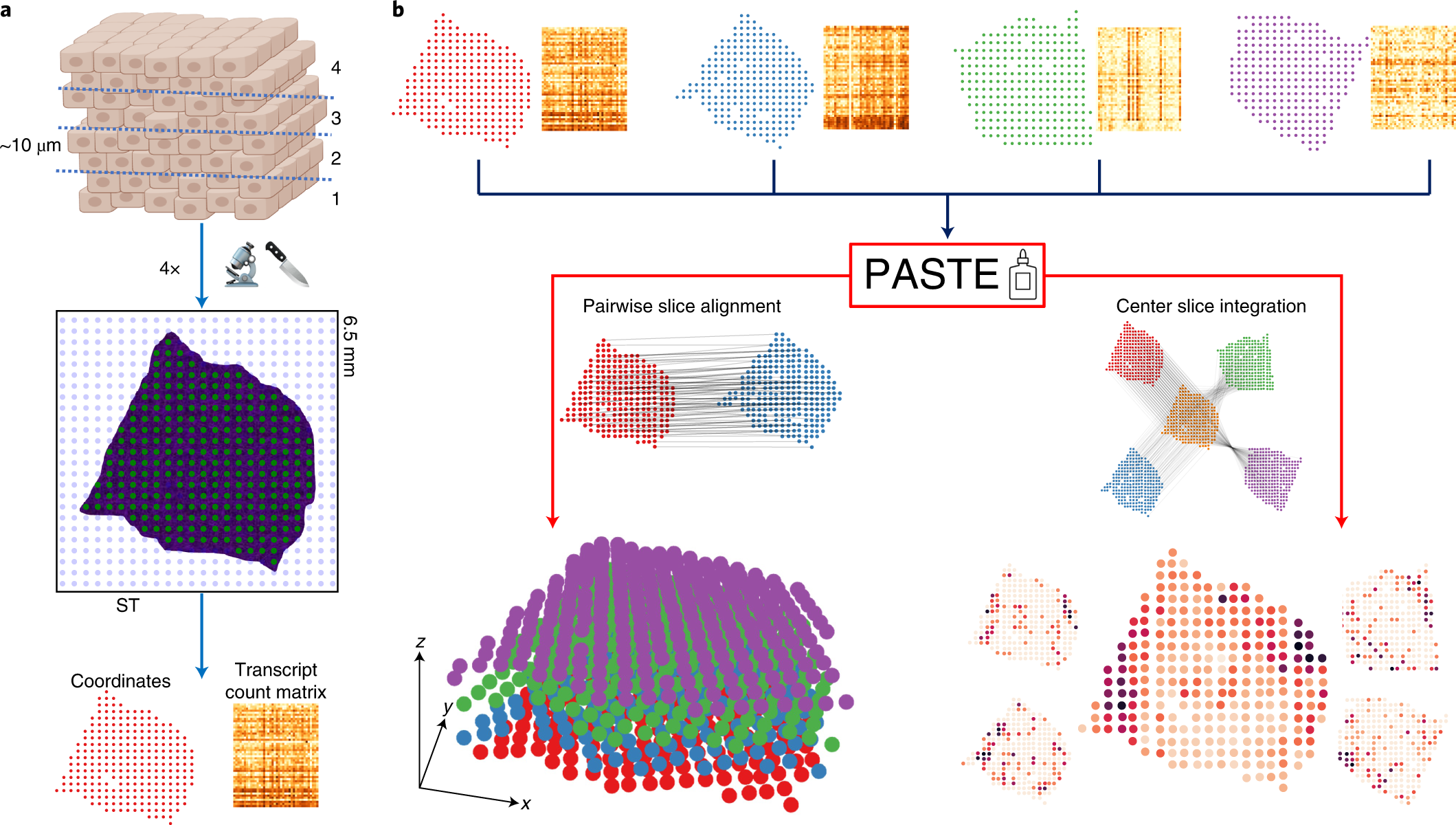
Alignment and integration of ST slices with PASTE
a, Each slice generated for an ST experiment is placed on a 2D grid of barcoded spots, and mRNA expression of each spot is measured along with the spatial coordinates of each spot. Only a fraction of spots (green) contain tissue cells, with other spots (blue) not covered by a tissue. This results in a transcript count matrix for the tissue spots together with their spatial coordinates. b, PASTE takes as input multiple ST slices consisting of spot expression matrices and spot spatial locations. In pairwise slice alignment mode, PASTE finds an optimal mapping between spots in one slice and spots in another slice while preserving the gene expression and the spatial distances of mapped spots. These mappings can then be used to reconstruct a stacked 3D alignment of the tissue by stacking slices on top of each other. In center slice integration mode, PASTE infers a ‘center’ slice consisting of a low rank expression matrix and a collection of mappings from the spots of the center slice to the spots of each input slice. The inferred center slice generally has lower sparsity and lower variance than the individual ST slices.
Zeira R, Land M, Strzalkowski A, Raphael BJ. (2022) Alignment and integration of spatial transcriptomics data. Nat Methods 19(5):567-575. [abstract]