With the advent of single cell/nucleus RNA sequencing (sc/snRNA-seq), the field of cell phenotyping is now a data-driven exercise providing statistical evidence to support cell type/state categorization. However, the task of classifying cells into specific, well-defined categories with the empirical data provided by sc/snRNA-seq remains nontrivial due to the difficulty in determining specific differences between related cell types with close transcriptional similarities, resulting in challenges with matching cell types identified in separate experiments.
To investigate possible approaches to overcome these obstacles, Researchers at the University of California, San Diego explored the use of supervised machine learning methods—logistic regression, support vector machines, random forests, neural networks, and light gradient boosting machine (LightGBM)–as approaches to classify cell types using snRNA-seq datasets from human brain middle temporal gyrus (MTG) and human kidney. Classification accuracy was evaluated using an F-beta score weighted in favor of precision to account for technical artifacts of gene expression dropout. The researchers examined the impact of hyperparameter optimization and feature selection methods on F-beta score performance. They found that the best performing model for granular cell type classification in both datasets is a multinomial logistic regression classifier and that an effective feature selection step was the most influential factor in optimizing the performance of the machine learning pipelines.
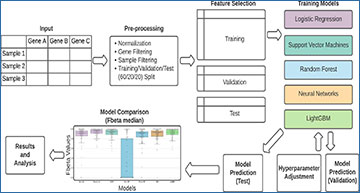
Overview of the machine learning pipeline
A count matrix undergoes pre-processing, including normalization and filtering. The data is randomly split into training (60%), validation (20%), and test (20%) sets independently for each cell type. The training sets are used to train the models. The validation set provides an initial test for accuracy of the trained models and is used to adjust the model’s hyperparameters. Once the hyperparameters are optimized, the test set is run through each model and the F-beta score distribution across all clusters is used for model comparison.
Availability – The code used to preprocess the data and run each model can be found in this GitHub repository: https://github.com/HuyGLe/neuronal-cell-type-classification.
Le H, Peng B, Uy J, Carrillo D, Zhang Y, Aevermann BD, et al. (2022) Machine learning for cell type classification from single nucleus RNA sequencing data. PLoS ONE 17(9): e0275070. [article]