The use of next-generation sequencing (NGS) has simplified the diagnosis of human genetic disorders. Since the mapping of the human genome, scientists have been able to detect diseases even at a “single gene” level. While DNA-sequencing provides information about the 3 billion bases of the human DNA, it fails to identify changes in the non-coding regions of the genome. These non-coding regions or intronic variations have recently been identified as key players in multiple disorders. Also, NGS techniques like whole-exome sequencing (WES) and whole-genome sequencing (WGS) are unable to determine the cause of several genetic disorders. This limits the overall diagnostic efficiency of NGS techniques.
RNA-sequencing, which is the analysis of the transcriptome, is a promising clinical diagnostic tool. It uses blood, fibroblasts, and muscle biopsies as samples and addresses the shortcomings of other sequencing methods. Data from RNA-sequencing provides detailed information on the sequence, structure, and quantity of specific RNA sequences. Experts believe that it has resulted in a nearly 15% diagnostic improvement by the systematic identification of RNA phenotypes!
Recently, researchers led by Dr. Holger Prokisch of the Technical University of Munich reviewed how RNA sequencing can improve the diagnostic power of NGS techniques in an article published in Pediatric Investigation on 5 March 2022.
According to Dr. Prokisch, “RNA-sequencing demonstrates success in prioritizing and detecting not only deleterious deep intronic variants but also coding variants affecting gene expression, which are often overlooked by WES and WGS.”
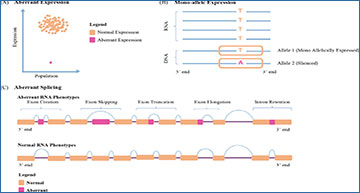
Three aberrant RNA phenotypes identified by RNA sequencing (RNA-seq)
(A) Aberrant expression, (B) Mono-allelic expression, and (C) Aberrant splicing versus normal slicing.
In their article, the researchers analyzed eight previous studies and found a 15% improvement in the performance of DNA-based sequencing when combined with RNA sequencing. The authors enlisted three possible reasons for abnormal RNA content in cells. They describe how “aberrant expression” causes a defect in the cell’s machinery during the process of transcription (converting DNA into RNA).
“Reduced expression of a disease gene often translates into low protein levels and indicates a genetic disorder,” explains Dr. Prokisch.
Next, they discuss “mono-allelic expression,” which results in the silencing of one allele or gene copy, and causes abnormal RNA profiles. Third, they explain how faulty processing of a newly formed RNA transcript, especially during “splicing” of non-protein coding regions, may be problematic. Some abnormal splicing events may result in sequences carrying rare mutations, resulting in abnormal RNA profiles.
RNA sequencing can easily identify these abnormal patterns, providing an edge in detecting genomic anomalies. The data from sequencing could be further used to improve the performance of in silico prediction tools. However, each technique has its own limitations. While DNA-sequencing is mainly tissue independent, RNA-sequencing relies heavily on tissue-specific expression.
“Different tissues or cell types represent a wide spectrum of splicing events and expression patterns, warranting caution when selecting and sampling blood and tissue for RNA-sequencing,” explains Dr. Prokisch.
Although RNA-sequencing is widely used around the world, its clinical application is still limited, highlighting the importance of incorporating additional NGS data, including proteomics, to correlate the RNA-sequencing findings, particularly for variants that do not have abnormal phenotypes. Moreover, improved analytical tools and guidelines for interpreting these variants must be incorporated into the bioinformatic guidelines for clinical use.
Dr. Prokisch concludes by saying, “If we can identify and leverage these abnormal phenotypes, identifying rare genetic diseases and their causes may become easier and more fulfilling in clinical practice.”
Source – PRNewswire
Peymani F, Farzeen A, Prokisch H. (2023) RNA sequencing role and application in clinical diagnostic. Pediatr Investig 6(1):29-35. [article]