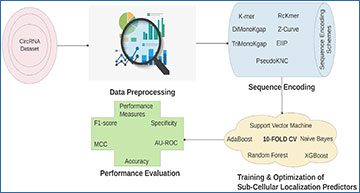
Circular ribonucleic acids (circRNAs) are novel non-coding RNAs that emanate from alternative splicing of precursor mRNA in reversed order across exons. Despite the abundant presence of circRNAs in human genes and their involvement in diverse physiological processes, the functionality of most circRNAs remains a mystery. Like other non-coding RNAs, sub-cellular localization knowledge of circRNAs has the aptitude to demystify the influence of circRNAs on protein synthesis, degradation, destination, their association with different diseases, and potential for drug development. To date, wet experimental approaches are being used to detect sub-cellular locations of circular RNAs. These approaches help to elucidate the role of circRNAs as protein scaffolds, RNA-binding protein (RBP) sponges, micro-RNA (miRNA) sponges, parental gene expression modifiers, alternative splicing regulators, and transcription regulators.
To complement wet-lab experiments, considering the progress made by machine learning approaches for the determination of sub-cellular localization of other non-coding RNAs, researchers at the German Research Center for Artificial Intelligence have developed a computational framework, Circ-LocNet, to precisely detect circRNA sub-cellular localization. Circ-LocNet performs comprehensive extrinsic evaluation of 7 residue frequency-based, residue order and frequency-based, and physio-chemical property-based sequence descriptors using the five most widely used machine learning classifiers. Further, it explores the performance impact of K-order sequence descriptor fusion where it ensembles similar as well dissimilar genres of statistical representation learning approaches to reap the combined benefits. Considering the diversity of statistical representation learning schemes, it assesses the performance of second-order, third-order, and going all the way up to seventh-order sequence descriptor fusion. A comprehensive empirical evaluation of Circ-LocNet over a newly developed benchmark dataset using different settings reveals that standalone residue frequency-based sequence descriptors and tree-based classifiers are more suitable to predict sub-cellular localization of circular RNAs. Further, K-order heterogeneous sequence descriptors fusion in combination with tree-based classifiers most accurately predict sub-cellular localization of circular RNAs. The researchers anticipate this study will act as a rich baseline and push the development of robust computational methodologies for the accurate sub-cellular localization determination of novel circRNAs.
CircLoc-Net: A Computational Framework for Sub-cellular Localization Prediction of circRNAs
Availability – Circ-LocNet is deployed as a very first circRNAs sub-cellular localization prediction platform at https://circ_rna_location_predictor.opendfki.de/.
Asim MN, Ibrahim MA, Malik MI, Dengel A, Ahmed S. (2022) Circ-LocNet: A Computational Framework for Circular RNA Sub-Cellular Localization Prediction. Int J Mol Sci 23(15), 8221. [article]